Gut Microbiome Diversity in European Honeybees (Apis mellifera L.) from La Union, Northern Luzon, Philippines
- PMID: 40003742
- PMCID: PMC11855267
- DOI: 10.3390/insects16020112
Gut Microbiome Diversity in European Honeybees (Apis mellifera L.) from La Union, Northern Luzon, Philippines
Abstract
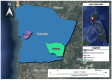
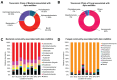
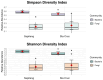
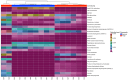
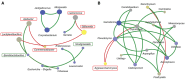
Insects often rely on symbiotic bacteria and fungi for various physiological processes, developmental stages, and defenses against parasites and diseases. Despite their significance, the associations between bacterial and fungal symbionts in Apis mellifera are not well studied, particularly in the Philippines. In this study, we collected A. mellifera from two different sites in the Municipality of Bacnotan, La Union, Philippines. A gut microbiome analysis was conducted using next-generation sequencing with the Illumina MiSeq platform. Bacterial and fungal community compositions were assessed using 16S rRNA and ITS gene sequences, respectively. Our findings confirm that adult worker bees of A. mellifera from the two locations possess distinct but comparably proportioned bacterial and fungal microbiomes. Key bacterial symbionts, including Lactobacillus, Bombilactobacillus, Bifidobacterium, Gilliamella, Snodgrassella, and Frischella, were identified. The fungal community was dominated by the yeasts Zygosaccharomyces and Priceomyces. Using the ENZYME nomenclature database and PICRUSt2 software version 2.5.2, a predicted functional enzyme analysis revealed the presence of β-glucosidase, catalase, glucose-6-phosphate dehydrogenase, glutathione transferase, and superoxide dismutase, which are involved in host defense, carbohydrate metabolism, and energy support. Additionally, we identified notable bacterial enzymes, including acetyl-CoA carboxylase and AMPs nucleosidase. Interestingly, the key bee symbionts were observed to have a negative correlation with other microbiota. These results provide a detailed characterization of the gut microbiota associated with A. mellifera in the Philippines and lay a foundation for further metagenomic studies of microbiomes in native or indigenous bee species in the region.
Keywords: Illumina MiSeq; Philippines; bacterial microbiota; fungal microbiota; honeybee functional microbiota; insects; metagenomics; next-generation sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Characterization of the Kenyan Honey Bee (Apis mellifera) Gut Microbiota: A First Look at Tropical and Sub-Saharan African Bee Associated Microbiomes.Microorganisms. 2020 Nov 3;8(11):1721. doi: 10.3390/microorganisms8111721. Microorganisms. 2020. PMID: 33153032 Free PMC article.
-
Genetic divergence and functional convergence of gut bacteria between the Eastern honey bee Apis cerana and the Western honey bee Apis mellifera.J Adv Res. 2021 Aug 10;37:19-31. doi: 10.1016/j.jare.2021.08.002. eCollection 2022 Mar. J Adv Res. 2021. PMID: 35499050 Free PMC article.
-
Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages.Microbiol Res. 2020 Jan;231:126370. doi: 10.1016/j.micres.2019.126370. Epub 2019 Nov 7. Microbiol Res. 2020. PMID: 31739261
-
Microbial ensemble in the hives: deciphering the intricate gut ecosystem of hive and forager bees of Apis mellifera.Mol Biol Rep. 2024 Feb 1;51(1):262. doi: 10.1007/s11033-024-09239-5. Mol Biol Rep. 2024. PMID: 38302671
-
Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis.Gut Microbes. 2013 Jan-Feb;4(1):60-5. doi: 10.4161/gmic.22517. Epub 2012 Oct 12. Gut Microbes. 2013. PMID: 23060052 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

