Mint3 as a Molecular Target Activated in the Early Stage of Hepatocarcinogenesis
- PMID: 40003897
- PMCID: PMC11855386
- DOI: 10.3390/ijms26041430
Mint3 as a Molecular Target Activated in the Early Stage of Hepatocarcinogenesis
Abstract
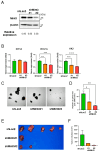
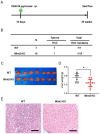
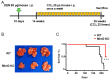
Mint3 enhances aerobic ATP production with subsequent nuclear translocation of hypoxia-inducible factor-1 (HIF-1) and activation of angiogenesis-related genes. It remains unclear if and when Mint3 is activated and whether it is involved in hepatocarcinogenesis. We explored the expression of Mint3 in surgically resected hepatocellular carcinoma (HCC) tissues. We evaluated the effects of Mint3 knockdown on spheroid formation capacity and subcutaneous tumor growth in immune-deficient mice. We used Mint3 knockout mice to evaluate the effects of chemically induced HCC development. Mint3 was overexpressed in well-differentiated HCC with the activation of HIF-1 target genes irrespective of the absence of hypervascularization. Mint3 knockdown ameliorated the expression of HIF-1 target genes in patient-derived HCC cell lines and suppressed spheroid formation. Mint3 knockdown further inhibited subcutaneous tumor formation in vivo in immune-deficient mice. Chemical HCC development induced by N-nitrosodiethylamine (DEN) or DEN/CCl4 was dramatically suppressed in Mint3 knockout mice compared to control mice. Mint3 plays a crucial role in early-stage HCC development before hypervascularization by activating HIF-1 target genes before the tumor becomes hypoxic. Mint3 is a molecular target that prevents HCC development in the early stages.
Keywords: hepatocellular carcinoma; hypoxia-inducible factor-1; munc18-1-interacting protein 3.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Llovet J.M., Willoughby C.E., Singal A.G., Greten T.F., Heikenwälder M., El-Serag H.B., Finn R.S., Friedman S.L. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: Pathogenesis and treatment. Nat. Rev. Gastroenterol. Hepatol. 2023;20:487–503. doi: 10.1038/s41575-023-00754-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

