Transcriptome-Wide Insights: Neonatal Lactose Intolerance Promotes Telomere Damage, Senescence, and Cardiomyopathy in Adult Rat Heart
- PMID: 40004050
- PMCID: PMC11855832
- DOI: 10.3390/ijms26041584
Transcriptome-Wide Insights: Neonatal Lactose Intolerance Promotes Telomere Damage, Senescence, and Cardiomyopathy in Adult Rat Heart
Abstract
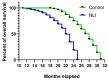
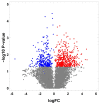
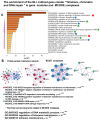
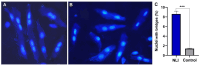
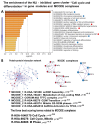
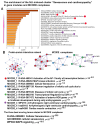
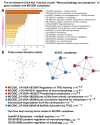
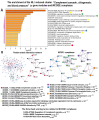
Cardiovascular diseases (CVD) are the primary cause of mortality globally. A significant aspect of CVD involves their association with aging and susceptibility to neonatal programming. These factors suggest that adverse conditions during neonatal development can disrupt cardiomyocyte differentiation, thereby leading to heart dysfunction. This study focuses on the long-term effects of inflammatory and oxidative stress due to neonatal lactose intolerance (NLI) on cardiomyocyte transcriptome and phenotype. Our recent bioinformatic study focused on toggle genes indicated that NLI correlates with the switch off of some genes in thyroid hormone, calcium, and antioxidant signaling pathways, alongside the switch-on/off genes involved in DNA damage response and inflammation. In the presented study, we evaluated cardiomyocyte ploidy in different regions of the left ventricle (LV), complemented by a transcriptomic analysis of genes with quantitative (gradual) difference in expression. Cytophotometric and morphologic analyses of LV cardiomyocytes identified hyperpolyploidy and bridges between nuclei suggesting telomere fusion. Transcriptomic profiling highlighted telomere damage, aging, and chromatin decompaction, along with the suppression of pathways governing muscle contraction and energy metabolism. Echocardiography revealed statistically significant LV dilation and a decrease in ejection fraction. The estimation of survival rates indicated that NLI shortened the median lifespan by approximately 18% (p < 0.0001) compared with the control. Altogether, these findings suggest that NLI may increase susceptibility to cardiovascular diseases by accelerating aging due to oxidative stress and increased telomere DNA damage, leading to hyperpolyploidization and reduced cardiac contractile function. Collectively, our data emphasize the importance of the early identification and management of neonatal inflammatory and metabolic stressors, such as NLI, to mitigate long-term cardiovascular risks.
Keywords: DNA damage; cardiomyocyte; cardiovascular disease; developmental programming; metabolic deprivation; neonatal lactose intolerance; polyploidy; senescence; telomere fusion.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures



References
-
- Kobelyatskaya A.A., Guvatova Z.G., Tkacheva O.N., Isaev F.I., Kungurtseva A.L., Vitebskaya A.V., Kudryavtseva A.V., Plokhova E.V., Machekhina L.V., Strazhesko I.D., et al. EchoAGE: Echocardiography-Based Neural Network Model Forecasting Heart Biological Age. Aging Dis. 2024;16 doi: 10.14336/AD.2024.0615. Published ahead of print . - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

