Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis
- PMID: 40004102
- PMCID: PMC11855550
- DOI: 10.3390/ijms26041638
Meta-Analysis of QTL Mapping and GWAS Reveal Candidate Genes for Heat Tolerance in Small Yellow Croaker, Larimichthys polyactis
Abstract
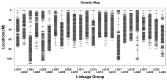
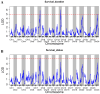
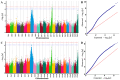
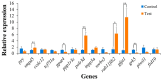
High temperatures present considerable challenges to global fish growth and production, yet the genetic basis of heat tolerance remains underexplored. This study combines quantitative trait locus (QTL) mapping and genome-wide association studies (GWAS) to examine the genetics of heat tolerance in Larimichthys polyactis. As a result, a genetic linkage map was constructed with 3237 bin markers spanning 24 linkage groups and totaling 1900.84 centimorgans, using genotyping-by-sequencing of a full-sib family comprising 120 progeny and their two parents. Based on this genetic linkage map, QTL mapping identified four QTLs associated with heat tolerance, which encompassed 18 single nucleotide polymorphisms and harbored 648 genes within the QTL intervals. The GWAS further disclosed 76 candidate genes related to heat tolerance, 56 of which overlapped with the QTL results. Enrichment analysis indicated that these genes are involved in immune response, development, lipid metabolism, and endocrine regulation. qPCR validation of 14 prioritized genes, which were simultaneously enriched in Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways, confirmed significant upregulation of smpd5, polr3d, rab11fip2, and gfpt1, along with downregulation of gpat4 and grk5 after 6 h of heat stress. These findings demonstrate their responsiveness to elevated high temperatures. This meta-analysis of QTL mapping and GWAS has successfully identified functional genes related to heat tolerance, enhancing understanding of the genetic architecture underlying this critical trait in L. polyactis. It also provides a molecular breeding tool to improve genetic traits associated with heat tolerance in cultured L. polyactis.
Keywords: GWAS; Larimichthys polyactis; QTL; genetic linkage map; heat tolerance.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
QTL Mapping-Based Identification of Visceral White-Nodules Disease Resistance Genes in Larimichthys polyactis.Int J Mol Sci. 2024 Oct 10;25(20):10872. doi: 10.3390/ijms252010872. Int J Mol Sci. 2024. PMID: 39456653 Free PMC article.
-
Constructing a High-Density Genetic Linkage Map for Large Yellow Croaker (Larimichthys crocea) and Mapping Resistance Trait Against Ciliate Parasite Cryptocaryon irritans.Mar Biotechnol (NY). 2019 Apr;21(2):262-275. doi: 10.1007/s10126-019-09878-x. Epub 2019 Feb 19. Mar Biotechnol (NY). 2019. PMID: 30783862
-
A first genetic linage map construction and QTL mapping for growth traits in Larimichthys polyactis.Sci Rep. 2020 Jul 15;10(1):11621. doi: 10.1038/s41598-020-68592-0. Sci Rep. 2020. PMID: 32669609 Free PMC article.
-
Underlying genetic architecture of resistance to mastitis in dairy cattle: A systematic review and gene prioritization analysis of genome-wide association studies.J Dairy Sci. 2023 Jan;106(1):323-351. doi: 10.3168/jds.2022-21923. Epub 2022 Nov 1. J Dairy Sci. 2023. PMID: 36333139
-
Human QTL linkage mapping.Genetica. 2009 Jun;136(2):333-40. doi: 10.1007/s10709-008-9305-3. Epub 2008 Jul 31. Genetica. 2009. PMID: 18668207 Free PMC article. Review.
References
-
- Bonfils C.J.W., Santer B.D., Fyfe J.C., Marvel K., Phillips T.J., Zimmerman S.R.H. Human influence on joint changes in temperature, rainfall and continental aridity. Nat. Clim. Chang. 2020;10:726–731. doi: 10.1038/s41558-020-0821-1. - DOI
-
- Pan L., Zhang L., Zhang Y., Xie S., Qin Q. Effects of acute high temperature stress on the growth, immune response, and antioxidant capacity of juvenile grass carp (Ctenopharyngodon idella) Aquac. Res. 2018;49:144–153.
-
- Li M., Chen L., Zeng C., Qin J.G., Chen H. Effects of high temperature stress on oxidative stress, immune response and heat shock protein gene expression in fish: A review. Aquac. Res. 2017;48:753–770. doi: 10.1111/are.12992. - DOI
-
- Xiong Y., Yang J., Jiang T., Liu H.B., Zhong X.M., Tang J.H. Early life history of the small yellow croaker (Larimichthys polyactis) in sandy ridges of the South Yellow Sea. Mar. Biol. Res. 2017;13:993–1002. doi: 10.1080/17451000.2017.1319067. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

