Evaluation of Pan-Cancer Immune Heterogeneity Based on DNA Methylation
- PMID: 40004489
- PMCID: PMC11855777
- DOI: 10.3390/genes16020160
Evaluation of Pan-Cancer Immune Heterogeneity Based on DNA Methylation
Abstract
Background/objectives: The heterogeneity of the tumor immune microenvironment is a key determinant of tumor oncogenesis. This study aims to evaluate the composition of seven immune cells across 5323 samples from 14 cancers using DNA methylation data.
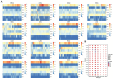
Methods: A deconvolution algorithm was proposed to estimate the composition of seven immune cells using 1256 immune cell population-specific methylation genes. Based on the immune infiltration features of seven immune cell fractions, 42 subtypes of 14 tumors (2-5 subtypes per tumor) were identified.
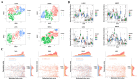
Results: Significant differences in immune cells between subtypes were revealed for each cancer. The study found that the methylation values of the selected specific sites correlated with gene expression in most tumor subtypes. Immune infiltration results were integrated with phenotypic data, including survival data and tumor stages, revealing significant correlations between immune infiltration and phenotypes in some tumors. Subtypes with high proportions of CD4+ T cells, CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes, neutrophils, and eosinophils were identified, with subtype counts of 9, 24, 22, 13, 19, 9, and 11, respectively. Additionally, 2412 differentially expressed genes between these subtypes and normal tissues were identified. Pathway enrichment analysis revealed that these genes were mainly enriched in pathways related to drug response and chemical carcinogens. Differences in ESTIMATE scores for subtypes of seven tumors and TIDE scores for eight tumors were also observed.
Conclusions: This study demonstrates the intra-tumor and inter-tumor immune heterogeneity of pan-cancer through DNA methylation analysis, providing assistance for tumor diagnosis.
Keywords: DNA methylation; clustering; deconvolution algorithm; pan-cancer; tumor immune microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Pan-cancer analysis reveals correlation between RAB3B expression and tumor heterogeneity, immune microenvironment, and prognosis in multiple cancers.Sci Rep. 2024 Apr 30;14(1):9881. doi: 10.1038/s41598-024-60581-x. Sci Rep. 2024. PMID: 38688977 Free PMC article.
-
Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data.BMC Bioinformatics. 2019 May 1;20(Suppl 7):195. doi: 10.1186/s12859-019-2734-4. BMC Bioinformatics. 2019. PMID: 31074374 Free PMC article.
-
Pan-cancer analysis of T-cell proliferation regulatory genes as potential immunotherapeutic targets.Aging (Albany NY). 2024 Jun 29;16(14):11224-11247. doi: 10.18632/aging.205977. Epub 2024 Jun 29. Aging (Albany NY). 2024. PMID: 39068665 Free PMC article.
-
Deciphering the nexus between the tumor immune microenvironment and DNA methylation in subgrouping estrogen receptor-positive breast cancer.Breast Cancer. 2021 Nov;28(6):1252-1260. doi: 10.1007/s12282-021-01262-9. Epub 2021 May 8. Breast Cancer. 2021. PMID: 33966175
-
Comprehensive scRNA-seq Analysis and Identification of CD8_+T Cell Related Gene Markers for Predicting Prognosis and Drug Resistance of Hepatocellular Carcinoma.Curr Med Chem. 2024;31(17):2414-2430. doi: 10.2174/0109298673274578231030065454. Curr Med Chem. 2024. PMID: 37936457
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

