Transcriptional Regulatory Network of the Embryonic Diapause Termination Process in Artemia
- PMID: 40004504
- PMCID: PMC11855619
- DOI: 10.3390/genes16020175
Transcriptional Regulatory Network of the Embryonic Diapause Termination Process in Artemia
Abstract
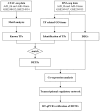
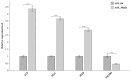
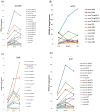
Artemia is a typical animal used for the study of the diapause mechanism. The research on the regulation mechanism of diapause mainly focuses on the occurrence and maintenance of diapause. There are few studies on the mechanism of embryonic pause termination (EDT), especially for its transcriptional regulation mechanism. This study integrated transcriptional regulatory data from ATAC-seq and gene expression data from RNA-seq to explore the transcriptional regulatory mechanisms involved in the EDT process. Through integrated analysis, four important transcription factors (TFs), SVP, MYC, RXR, and SMAD6, were found to play a role in the EDT process, in which SVP, MYC, and RXR were upregulated, while SMAD6 was downregulated in the EDT stage. Through co-expression analysis, a transcription regulatory network for these four TFs was constructed and the functions of the TFs were analyzed. The expression of the TFs was further verified by RT-qPCR. Through functional analysis, SVP was found to be predominantly involved in cell adhesion and signal transduction. MYC probably played a role in protein binding. RXR may function in the process of RNA binding and the transfer of phosphorus-containing groups. Smad6 regulated the signal transduction, cell adhesion, and oxidation-reduction processes. The expression of the key TFs was verified by RT-qPCR. The results of this work provide important clues for the mechanism of transcriptional regulation in the EDT process of Artemia.
Keywords: Artemia; diapause; embryonic pause termination; regulatory network; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Integrating ATAC-Seq and RNA-Seq Reveals the Signal Regulation Involved in the Artemia Embryonic Reactivation Process.Genes (Basel). 2024 Aug 16;15(8):1083. doi: 10.3390/genes15081083. Genes (Basel). 2024. PMID: 39202442 Free PMC article.
-
The Potential Roles of the Apoptosis-Related Protein PDRG1 in Diapause Embryo Restarting of Artemia sinica.Int J Mol Sci. 2018 Jan 2;19(1):126. doi: 10.3390/ijms19010126. Int J Mol Sci. 2018. PMID: 29301330 Free PMC article.
-
Signaling Transduction Pathways and G-Protein-Coupled Receptors in Different Stages of the Embryonic Diapause Termination Process in Artemia.Curr Issues Mol Biol. 2024 Apr 20;46(4):3676-3693. doi: 10.3390/cimb46040229. Curr Issues Mol Biol. 2024. PMID: 38666959 Free PMC article.
-
Stress tolerance during diapause and quiescence of the brine shrimp, Artemia.Cell Stress Chaperones. 2016 Jan;21(1):9-18. doi: 10.1007/s12192-015-0635-7. Epub 2015 Sep 3. Cell Stress Chaperones. 2016. PMID: 26334984 Free PMC article. Review.
-
Molecular chaperones, stress resistance and development in Artemia franciscana.Semin Cell Dev Biol. 2003 Oct;14(5):251-8. doi: 10.1016/j.semcdb.2003.09.019. Semin Cell Dev Biol. 2003. PMID: 14986854 Review.
Cited by
-
Using diapause as a platform to understand the biology of dormancy.Open Biol. 2025 Aug;15(8):250104. doi: 10.1098/rsob.250104. Epub 2025 Aug 20. Open Biol. 2025. PMID: 40829645 Free PMC article. Review.
References
-
- Qiu Z., MacRae T.H. A Molecular Overview of Diapause in Embryos of the Crustacean, Artemia franciscana. Volume 21 Springer; Berlin/Heidelberg, Germany: 2010.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

