The Impact of the Competition on miRNA, Proteins, and Metabolites in the Blood Exosomes of the Yili Horse
- PMID: 40004554
- PMCID: PMC11855450
- DOI: 10.3390/genes16020224
The Impact of the Competition on miRNA, Proteins, and Metabolites in the Blood Exosomes of the Yili Horse
Abstract
Purpose: Horse racing may cause stress-induced physiological changes and tissue damage in horses, but the changes in miRNA expression, protein expression, and metabolic substances in the plasma exosomes of the Yili horse after racing are still unclear. This study detected miRNA, protein expression, and metabolic substances in the plasma exosomes of Yili horses before and after competition, providing new insights for post-race recovery and care of Yili horses.
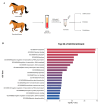
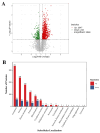
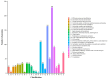
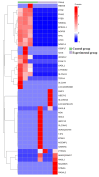
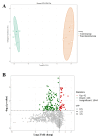
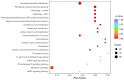
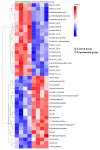
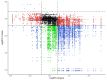
Method: Eight three-year-old Yili horses that had undergone training were selected as the research subjects, with four horses that had not competed as the control group and four horses that had participated in the competition for half an hour as the training group. Extract whole blood and separate plasma from two groups of horses, and then extract plasma exosomes; MiRNAs, proteins, and metabolites in extracellular vesicles were detected and analyzed using miRNAomics, proteomics, and metabolomics. P Result: After the competition, the levels of miRNAs related to the cytoplasm and nucleus in Yili horse plasma exosomes increased, and miRNAs related to the transcription and transcriptional regulation of biological processes significantly increased. The levels of proteins related to the cytoplasm and nucleus also increased, and the levels of proteins related to cell signaling function increased, carbohydrates and their metabolites were significantly reduced.
Conclusions: The competition process causes significant changes in the miRNA, proteomics, and metabolomics of plasma exosomes in the Yili horses, which are mainly related to metabolic regulation.
Keywords: Yili horse; competition; extracellular vesicles; metabolomics; miRNAomics; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Effect of β-alanine on the athletic performance and blood amino acid metabolism of speed-racing Yili horses.Front Vet Sci. 2024 Feb 28;11:1339940. doi: 10.3389/fvets.2024.1339940. eCollection 2024. Front Vet Sci. 2024. PMID: 38482164 Free PMC article.
-
Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses.Int J Mol Sci. 2025 Mar 8;26(6):2426. doi: 10.3390/ijms26062426. Int J Mol Sci. 2025. PMID: 40141070 Free PMC article.
-
Estimation of genetic parameters of body conformation and racing performance traits in Yili horses.J Equine Vet Sci. 2025 Mar;146:105378. doi: 10.1016/j.jevs.2025.105378. Epub 2025 Feb 6. J Equine Vet Sci. 2025. PMID: 39922246
-
Exosomes provide unappreciated carrier effects that assist transfers of their miRNAs to targeted cells; I. They are 'The Elephant in the Room'.RNA Biol. 2021 Nov;18(11):2038-2053. doi: 10.1080/15476286.2021.1885189. Epub 2021 May 4. RNA Biol. 2021. PMID: 33944671 Free PMC article. Review.
-
Metabolomics in equine sport and exercise.J Anim Physiol Anim Nutr (Berl). 2021 Jan;105(1):140-148. doi: 10.1111/jpn.13384. Epub 2020 Jun 8. J Anim Physiol Anim Nutr (Berl). 2021. PMID: 32511844 Review.
Cited by
-
Extracellular Vesicles in Sport Horses: Potential Biomarkers and Modulators of Exercise Adaptation and Therapeutics.Int J Mol Sci. 2025 May 3;26(9):4359. doi: 10.3390/ijms26094359. Int J Mol Sci. 2025. PMID: 40362597 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

