Towards Completion of the "Periodic Table" of Di-2-Pyridyl Ketoxime
- PMID: 40005105
- PMCID: PMC11858227
- DOI: 10.3390/molecules30040791
Towards Completion of the "Periodic Table" of Di-2-Pyridyl Ketoxime
Abstract
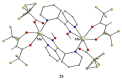
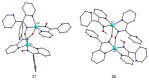
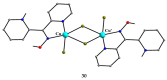
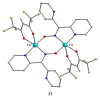
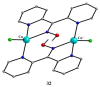
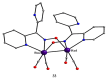
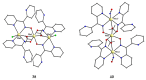
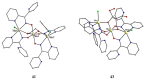
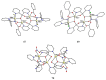
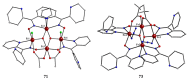
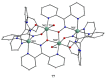
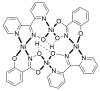
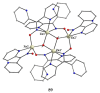
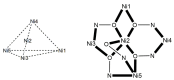
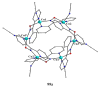
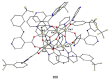
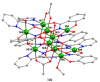
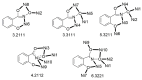
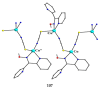
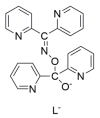
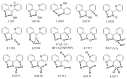
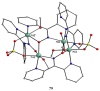
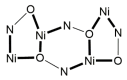
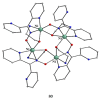
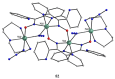
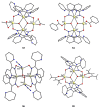
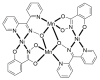
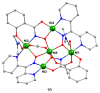
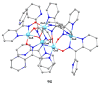
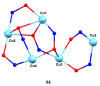
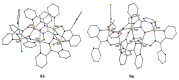
The oxime group is important in organic and inorganic chemistry. In most cases, this group is part of an organic molecule possessing one or more donor sites capable of forming bonds to metal ions. One family of such compounds is the group of 2-pyridyl (aldo)ketoximes. Metal complexes of 2-pyridyl oximes continue to attract the intense interest of many inorganic chemistry groups around the world for a variety of reasons, including their interesting structures, physical and biological properties, and applications. A unique member of 2-pyridyl ketoximes is di-2-pyridyl ketoxime (dpkoxH), which contains two 2-pyridyl groups and an oxime functionality that can be easily deprotonated giving the deprotonated ligand (dpkox-). The extra 2-pyridyl site confers a remarkable flexibility resulting in metal complexes with exciting structural and reactivity features. Our and other research groups have prepared and characterized many metal complexes of dpkoxH and dpkox- over the past 30 years or so. This work is an attempt to build a "periodic table" of dpkoxH, which is near completion. The filled spaces of this "periodic table" contain metal ions whose dpkoxH/dpkox- complexes have been structurally characterized. This work reviews comprehensively the to-date published coordination chemistry of dpkoxH with emphasis on the syntheses, reactivity, relationship to metallacrown chemistry, structures, and properties of the metal complexes; selected unpublished results from our group are also reported. The sixteen coordination modes adopted by dpkoxH and dpkox- have provided access to monomeric and dimeric complexes, trinuclear, tetranuclear, pentanuclear, hexanuclear, heptanuclear, enneanuclear, and decanuclear clusters, as well as to a small number of 1D coordination polymers. With few exceptions ({MIILnIII2} and {NiII2MnIII2}; M = Ni, Cu, Pd, and Ln = lanthanoid), most complexes are homometallic. The metals whose ions have yielded complexes with dpkoxH and dpkox- are Cr, Mn, Fe, Co, Ni, Cu, Zn, Ru, Rh, Pd, Ag, Cd, Re, Os, Ir, Au, Hg, lanthanoids (mainly Pr and Nd), and U. Most metal complexes are homovalent, but some mixed-valence Mn, Fe, and Co compounds have been studied. Metal ion-assisted/promoted transformations of dpkoxH, i.e., reactivity patterns of the coordinated ligand, are also critically discussed. Some perspectives concerning the coordination chemistry of dpkoxH and research work for the future are outlined.
Keywords: coordination chemistry; di-2-pyridyl ketoxime’s metal complexes; magnetic properties; reactivity; structures; synthesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures















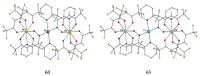
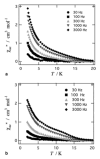
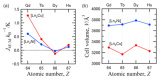
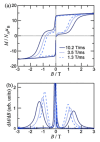












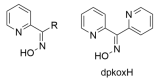


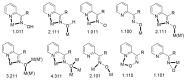



















Similar articles
-
Reactions of Cadmium(II) Halides and Di-2-Pyridyl Ketone Oxime: One-Dimensional Coordination Polymers.Molecules. 2024 Jan 19;29(2):509. doi: 10.3390/molecules29020509. Molecules. 2024. PMID: 38276587 Free PMC article.
-
Binding of oxime group to uranyl ion.Dalton Trans. 2016 May 31;45(22):9307-19. doi: 10.1039/c6dt01293k. Dalton Trans. 2016. PMID: 27184620
-
A structure-based analysis of the vibrational spectra of nitrosyl ligands in transition-metal coordination complexes and clusters.Spectrochim Acta A Mol Biomol Spectrosc. 2011 Jan;78(1):7-28. doi: 10.1016/j.saa.2010.08.001. Epub 2010 Aug 17. Spectrochim Acta A Mol Biomol Spectrosc. 2011. PMID: 21123107
-
Azide groups in higher oxidation state manganese cluster chemistry: from structural aesthetics to single-molecule magnets.Inorg Chem. 2009 Apr 20;48(8):3308-22. doi: 10.1021/ic801217j. Inorg Chem. 2009. PMID: 19364123 Review.
-
Coordination Chemistry of Nucleotides and Antivirally Active Acyclic Nucleoside Phosphonates, including Mechanistic Considerations.Molecules. 2022 Apr 19;27(9):2625. doi: 10.3390/molecules27092625. Molecules. 2022. PMID: 35565975 Free PMC article. Review.
References
-
- March J. Advanced Organic Chemistry. 4th ed. Wiley; New York, NY, USA: 1992. pp. 73, 128, 367, 388, 389, 405, 406, 894, 918, 934, 943, 1038–1040, 1154, 1199.
-
- Coxall R.A., Harris S.G., Henderson D.K., Parsons S., Tasker P.A., Winpenny R.E.P. Inter-Ligand Reactions: In Situ Formation of New Polydentate Ligands. J. Chem. Soc. Dalton Trans. 2000:2349–2356. doi: 10.1039/b001404o. - DOI
-
- Milios C.J., Stamatatos T.C., Perlepes S.P. The coordination chemistry of pyridyl oximes. Polyhedron. 2006;25:134–194. doi: 10.1016/j.poly.2005.07.022. - DOI
-
- Tschugaeff L. Ueber ein neues, empfindliches reagens out nickel. Ber. Dtsch. Chem. Ges. 1885;38:2520–2522. doi: 10.1002/cber.19050380317. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

