Differentially Expressed miRNA Profiles in Serum-Derived Exosomes from Cattle Infected with Lumpy Skin Disease Virus
- PMID: 40005551
- PMCID: PMC11858326
- DOI: 10.3390/pathogens14020176
Differentially Expressed miRNA Profiles in Serum-Derived Exosomes from Cattle Infected with Lumpy Skin Disease Virus
Abstract
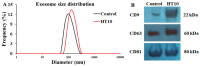
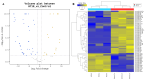
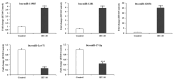
Exosomal miRNAs from individual cells are crucial in regulating the immune response to infectious diseases. In this study, we performed small RNA sequencing (small RNA-seq) analysis to identify the expressed and associated exosomal miRNAs in the serum of cattle infected with lumpy skin disease virus (LSDV). Cattle were infected with a 106.5 TCID50/mL LSDV Vietnam/HaTinh/CX01 (HT10) strain and exosomal miRNA expression in the serum of infected cattle was analyzed using small RNA sequencing (small RNA-seq). We identified 59 differentially expressed (DE) miRNAs in LSDV-infected cattle compared to uninfected controls, including 18 upregulated and 41 downregulated miRNAs. These 59 miRNAs were used to predict 7656 target genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that the target genes were enriched in several biological processes and pathways associated with viral replication, immune response, virus-host interactions, and signal transduction. Additionally, we identified 708 potentially novel cattle miRNAs corresponding to 710 genomic loci. The transcription levels of five miRNA genes (bta-miR-11985, bta-miR-1281, bta-miR-12034, bta-miR-let-7i, and bta-miR-17-5p) were validated using reverse transcription quantitative real-time PCR, showing consistency with the small RNA-seq results. Overall, these findings provide significant insights into the immune and protective responses during LSDV infection in cattle, offering valuable information on identifying new biomarkers and understanding the pathogenesis of LSDV.
Keywords: exosome; lumpy skin disease; miRNA; pathway; small RNA-seq.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- WOAH . OIE Terrestrial Manual 2021. WOAH; Paris, France: 2021. Infection with Lumpy Skin Disease Virus. Chapter 11.9.
-
- Casal J., Allepuz A., Miteva A., Pite L., Tabakovsky B., Terzievski D., Alexandrov T., Beltran-Alcrudo D. Economic cost of lumpy skin disease outbreaks in three Balkan countries: Albania, Bulgaria and the Former Yugoslav Republic of Macedonia (2016–2017) Transbound. Emerg. Dis. 2018;65:1680–1688. doi: 10.1111/tbed.12926. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

