Distinct Intraspecies Variation of Cutibacterium acnes and Staphylococcus epidermidis in Acne Vulgaris and Healthy Skin
- PMID: 40005665
- PMCID: PMC11858094
- DOI: 10.3390/microorganisms13020299
Distinct Intraspecies Variation of Cutibacterium acnes and Staphylococcus epidermidis in Acne Vulgaris and Healthy Skin
Abstract
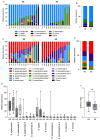
Human skin hosts a diverse array of microorganisms that contribute to its health. Key players in the facial skin microbiome include Cutibacterium acnes and staphylococci, whose colonization patterns may influence dermatological conditions like acne vulgaris. This study examined the facial microbiome composition of 29 individuals, including 14 with moderate to severe acne and 15 with healthy skin, using single locus sequence typing (SLST) amplicon sequencing. The results showed a shift in the relative abundances of C. acnes phylotypes: SLST types A, C, and F were increased in acne, while types H, K, and L were reduced compared to healthy skin. Among staphylococci, the relative abundance of S. epidermidis, S. capitis, and S. saphrophyticus increased in acne, while S. saccharolyticus and S. hominis decreased. The amplicon sequencing approach could also identify a population shift of S. epidermidis: a specific S. epidermidis phylogenetic lineage (type 3) was reduced in acne, while two abundant lineages (types 1 and 2) were elevated. These findings suggest that distinct phylogenetic lineages of both C. acnes and S. epidermidis are linked to healthy versus diseased skin, highlighting a potential role for both microorganisms in disease prevention and aggravation, respectively.
Keywords: Cutibacterium acnes; Staphylococcus; Staphylococcus epidermidis; acne vulgaris; amplicon-based next-generation sequencing; skin microbiome.
Conflict of interest statement
T.H., J.G., S.G., and J.H. are employees at Beiersdorf AG. The other authors declare no conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources

