Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application
- PMID: 40005754
- PMCID: PMC11858486
- DOI: 10.3390/microorganisms13020388
Genomic Insights into Probiotic Lactococcus lactis T-21, a Wild Plant-Associated Lactic Acid Bacterium, and Its Preliminary Clinical Safety for Human Application
Abstract
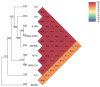
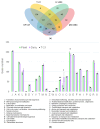
Lactococcus lactis T-21 is a lactic acid bacterium isolated from wild cranberries in Japan that demonstrates significant immunomodulatory properties and has been incorporated into commercial health products. However, probiogenomic analyses specific to T-21 have remained largely unexplored. This study performed a thorough genomic characterisation of T-21 and evaluated its safety in initial clinical trials. Genomic analysis revealed substantial genetic diversity and metabolic capabilities, including enhanced fermentative potential demonstrated by its ability to metabolise a wide range of plant-derived carbohydrates, and genetic determinants associated with exopolysaccharide biosynthesis and nisin production, distinguishing T-21 from domesticated dairy strains. These attributes, reflective of its wild plant origin, may contribute to its metabolic versatility and unique probiotic functionalities. A preliminary clinical trial assessing the safety of T-21-fermented milk in healthy Japanese adults indicated no significant adverse outcomes, corroborating its safety for human consumption. Together, these findings support the feasibility of utilising non-dairy, wild plant-origin strains in dairy fermentation processes as probiotics. This study expands our understanding of the genomic basis for T-21's probiotic potential and lays the groundwork for further investigations into its functional mechanisms and potential applications in promoting human health.
Keywords: Lactococcus lactis; clinical safety; comparative genomics; human health application; immunomodulatory properties; plant-associated LAB; probiotics.
Conflict of interest statement
Authors Masanori Fukao, Keisuke Tagawa and Shuichi Segawa were employed by the company Nissin York Co., Ltd. Authors Yosuke Sunada, Kazuya Uehara, Takuya Sugimoto were employed by the company Nissin Foods Holdings Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare that this study received funding from Nissin York Co., Ltd. and Nissin Foods Holdings. The funder had the following involvement with the study: Takeshi Zendo and Jiro Nakayama benefitted from a research grant from Nissin York. The test products used in this study were supplied by Nissin York for the clinical trials. Nissin York delegated the execution of this research to M&I Science, which conducted the study in collaboration with AMC Nishi-Umeda Clinic.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

