Could the Microbial Profiling of Normal Pancreatic Tissue from Healthy Organ Donors Contribute to Understanding the Intratumoral Microbiota Signature in Pancreatic Ductal Adenocarcinoma?
- PMID: 40005817
- PMCID: PMC11858623
- DOI: 10.3390/microorganisms13020452
Could the Microbial Profiling of Normal Pancreatic Tissue from Healthy Organ Donors Contribute to Understanding the Intratumoral Microbiota Signature in Pancreatic Ductal Adenocarcinoma?
Abstract
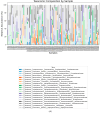
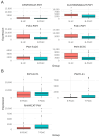
Pancreatic ductal adenocarcinoma (PDAC) is associated with intratumoral microbiota changes. However, defining the normal pancreatic microbial composition remains a challenge. Herein, we tested the hypothesis that the microbial profiling of normal pancreatic tissue from healthy organ donors (HC) could help in determining the signature of microbiota in PDAC. Matched pairs of tumor and normal tissues from PDAC patients (n = 32) and normal pancreatic tissues from HC (n = 17) were analyzed by 16S rRNA gene sequencing. Dissimilarities in all the beta metrics emerged in both normal samples and tumor samples, compared to HC (Bray-Curtis dissimilarity and Jaccard distance: p = 0.002; weighted UniFrac distances: p = 0.42 and p = 0.012, respectively; unweighted UniFrac distance: p = 0.009); a trend toward a lower Faith's phylogenetic distance was found at the tumor level vs. HC (p = 0.08). Within PDAC, a lower Faith's phylogenetic distance (p = 0.003) and a significant unweighted UniFrac distance (p = 0.024) were observed in tumor samples vs. normal samples. We noted the presence of a decreased abundance of bacteria with potential beneficial effects (Jeotgalicoccus) and anticancer activity (Acinetobacter_guillouiae) in PDAC vs. HC; bacteria involved in immune homeostasis and suppression of tumor progression (Streptococcus_salivarius, Sphingomonas) were reduced, and those implicated in tumor initiation and development (Methylobacterium-Methylorubrum, g_Delftia) were enhanced in tumor samples vs. normal samples. Metagenomic functions involved in fatty acid synthesis were reduced in normal samples compared to HC, while peptidoglycan biosynthesis IV and L-rhamnose degradation were more abundant in tumor samples vs. normal samples. Future prospective studies on larger populations, also including patients in advanced tumor stages and considering all potential existing confounding factors, as well as further functional investigations, are needed to prove the role of microbiota-mediated pathogenicity in PDAC.
Keywords: 16S rRNA gene sequencing; carcinogenesis; intratumoral bacteria; microbiota; pancreatic ductal adenocarcinoma; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

