Administration of a Recombinant Fusion Protein of IFN-γ and CD154 Inhibited the Infection of Chicks with Salmonella enterica
- PMID: 40005871
- PMCID: PMC11861687
- DOI: 10.3390/vetsci12020112
Administration of a Recombinant Fusion Protein of IFN-γ and CD154 Inhibited the Infection of Chicks with Salmonella enterica
Abstract
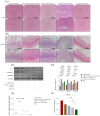
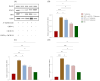
The cytokines IFN-γ and CD154 have been well established, and they play pivotal roles in immune protection against Salmonella in mice, but their effects and specific mechanisms in Salmonella-infected chickens are less understood. In this study, we conducted animal experiments to screen the highly immunoprotective chIFN-γ-chCD154 fusion protein compared with single protein chIFN-γ or chCD154 in white Leghorn chickens. The results showed that compared with separate pretreatments with chIFN-γ and chCD154, the fusion protein, chIFN-γ-chCD154, synergistically increased survival of infected chickens, reduced bacterial load in feces and organs, and attenuated pathological damage to the liver and cecum. Pretreatment with chIFN-γ-chCD154 also increased humoral immune responses, expression of the tight junction proteins zo-1, occludin, and claudin-1, and the relative abundance of Enterococcus_cecorum, Lactobacillus_helveticus, and Lactobacillus_agilis, which protect against intestinal inflammation. Compared with single protein pretreatment, chIFN-γ-chCD154 significantly upregulated STAT1, IRF1, and GBP1 in infected chickens while decreasing mRNA expression of TLR4, MyD88, NF-κB, TNF-α, IL-6, and IL-1β. In summary, damage to the cecal epithelial barrier and the inflammation induced by S. typhimurium infection was alleviated by chIFN-γ-chCD154 pretreatment through a mechanism involving the TLR4/MyD88/NF-κB and IFN-γ/STAT/IRF1/GBP1 pathways.
Keywords: CD154; IFN-γ; Salmonella Typhimurium; cecum; inflammation.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Oral administration of live attenuated Salmonella enterica serovar Typhimurium expressing chicken interferon-α alleviates clinical signs caused by respiratory infection with avian influenza virus H9N2.Vet Microbiol. 2011 Dec 29;154(1-2):140-51. doi: 10.1016/j.vetmic.2011.06.034. Epub 2011 Jul 2. Vet Microbiol. 2011. PMID: 21764226
-
Eugenol alleviates Salmonella Typhimurium-infected cecal injury by modulating cecal flora and tight junctions accompanied by suppressing inflammation.Microb Pathog. 2023 Jun;179:106113. doi: 10.1016/j.micpath.2023.106113. Epub 2023 Apr 14. Microb Pathog. 2023. PMID: 37062493
-
Enhancement of the anti-Salmonella immune response in CD154-deficient mice by an attenuated, IFN-γ-expressing, strain of Salmonella enterica serovar Typhimurium.Microb Pathog. 2012 Jun;52(6):326-35. doi: 10.1016/j.micpath.2012.03.002. Epub 2012 Mar 13. Microb Pathog. 2012. PMID: 22445817
-
Effects of multi-strain probiotic supplementation on intestinal microbiota, tight junctions, and inflammation in young broiler chickens challenged with Salmonella enterica subsp. enterica.Asian-Australas J Anim Sci. 2020 Nov;33(11):1797-1808. doi: 10.5713/ajas.19.0427. Epub 2019 Nov 12. Asian-Australas J Anim Sci. 2020. PMID: 32054193 Free PMC article.
-
Vaccination of chickens with recombinant Salmonella expressing M2e and CD154 epitopes increases protection and decreases viral shedding after low pathogenic avian influenza challenge.Poult Sci. 2009 Nov;88(11):2244-52. doi: 10.3382/ps.2009-00251. Poult Sci. 2009. PMID: 19834072 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

