Design, Synthesis, and Antiproliferative Activity of Novel Indole/1,2,4-Triazole Hybrids as Tubulin Polymerization Inhibitors
- PMID: 40006087
- PMCID: PMC11859928
- DOI: 10.3390/ph18020275
Design, Synthesis, and Antiproliferative Activity of Novel Indole/1,2,4-Triazole Hybrids as Tubulin Polymerization Inhibitors
Abstract
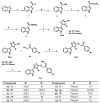
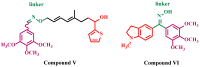
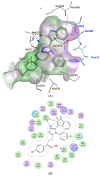
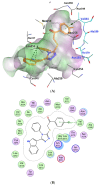
Background/Objectives: New indole/1,2,4-triazole hybrids were synthesized and tested for antiproliferative activity against the NCI 60 cell line as tubulin polymerization inhibitors. Methods: All final compounds, 6a-j and 7a-j were evaluated at a single concentration of 10 µM against a panel of sixty cancer cell lines. Results: Compounds 7a-j, featuring the NO-releasing oxime moiety, exhibited superior anticancer activity to their precursor ketones 6a-j across all tested cancer cell lines. Compounds 6h, 7h, 7i, and 7j were chosen for five-dose evaluations against a comprehensive array of 60 human tumor cell lines. The data showed that all tested compounds had significant anticancer activity throughout the nine tumor subpanels studied, with selectivity ratios ranging from 0.52 to 2.29 at the GI50 level. Compounds 7h and 7j showed substantial anticancer effectiveness against most cell lines across nine subpanels, with GI50 values ranging from 1.85 to 5.76 µM and 2.45 to 5.23 µM. Compounds 6h, 7h, 7i, and 7j were assessed for their inhibitory effects on tubulin polymerization. Conclusions: The results showed that compound 7i, an oxime-based derivative, was the most effective at blocking tubulin, with an IC50 value of 3.03 ± 0.11 µM. This was compared to the standard drug CA-4, which had an IC50 value of 8.33 ± 0.29 µM. Additionally, cell cycle analysis and apoptosis assays were performed for compound 7i. Molecular computational investigations have been performed to examine the binding mode of the most effective compounds to the target enzyme.
Keywords: CA-4; NCI; anticancer; cancer; colchicine; tubulin.
Conflict of interest statement
The authors state that they have no known competing financial interests or relationships that could have affected the work described in this study.
Figures






Similar articles
-
Synthesis and Preclinical Evaluation of Indole Triazole Conjugates as Microtubule Targeting Agents that are Effective against MCF-7 Breast Cancer Cell Lines.Anticancer Agents Med Chem. 2021;21(8):1047-1055. doi: 10.2174/1871520620666200925102940. Anticancer Agents Med Chem. 2021. PMID: 32981511
-
Discovery and optimization of 2,3-diaryl-1,3-thiazolidin-4-one-based derivatives as potent and selective cytotoxic agents with anti-inflammatory activity.Eur J Med Chem. 2023 Nov 5;259:115712. doi: 10.1016/j.ejmech.2023.115712. Epub 2023 Aug 6. Eur J Med Chem. 2023. PMID: 37567059
-
Design and discovery of new antiproliferative 1,2,4-triazin-3(2H)-ones as tubulin polymerization inhibitors targeting colchicine binding site.Bioorg Chem. 2021 Jul;112:104965. doi: 10.1016/j.bioorg.2021.104965. Epub 2021 May 5. Bioorg Chem. 2021. PMID: 34020238
-
Synthesis, anticancer activity and molecular modeling studies of 1,2,4-triazole derivatives as EGFR inhibitors.Eur J Med Chem. 2018 Aug 5;156:774-789. doi: 10.1016/j.ejmech.2018.07.024. Epub 2018 Jul 10. Eur J Med Chem. 2018. PMID: 30055463
-
New 1,2,3-Triazole/1,2,4-triazole Hybrids as Aromatase Inhibitors: Design, Synthesis, and Apoptotic Antiproliferative Activity.Molecules. 2023 Oct 14;28(20):7092. doi: 10.3390/molecules28207092. Molecules. 2023. PMID: 37894571 Free PMC article.
Cited by
-
New 1,2,4-Triazole Derivatives with a N-Mannich Base Structure Based on a 4,6-Dimethylpyridine Scaffold as Anticancer Agents: Design, Synthesis, Biological Evaluation, and Molecular Modeling.Int J Mol Sci. 2025 Jul 8;26(14):6572. doi: 10.3390/ijms26146572. Int J Mol Sci. 2025. PMID: 40724822 Free PMC article.
-
Synthesis and multi-target antiproliferative evaluation of novel 1,2,4-triazole-3-thione analogues against breast cancer: in silico and in vitro mechanistic insights.RSC Adv. 2025 Jul 14;15(30):24769-24790. doi: 10.1039/d5ra02512e. eCollection 2025 Jul 10. RSC Adv. 2025. PMID: 40661216 Free PMC article.
-
Design, Synthesis, and Pharmacological Activity of the N-(4-(1H-1,2,4-Triazol-1-yl)phenyl)-substituted-amide Derivatives.Molecules. 2025 Aug 18;30(16):3400. doi: 10.3390/molecules30163400. Molecules. 2025. PMID: 40871553 Free PMC article.
References
-
- Ruddon R.W. Cancer Biology. Oxford University Press; Oxford, UK: 2007.
LinkOut - more resources
Full Text Sources

