Evaluation of Genomic Surveillance of SARS-CoV-2 Virus Isolates and Comparison of Mutational Spectrum of Variants in Bangladesh
- PMID: 40006937
- PMCID: PMC11860708
- DOI: 10.3390/v17020182
Evaluation of Genomic Surveillance of SARS-CoV-2 Virus Isolates and Comparison of Mutational Spectrum of Variants in Bangladesh
Abstract
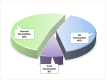
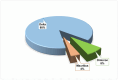
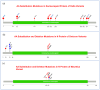
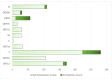
The SARS-CoV-2-induced disease, COVID-19, remains a worldwide public health concern due to its high rate of transmission, even in vaccinated and previously infected people. In the endemic state, it continues to cause significant pathology. To elu- cidate the viral mutational changes and screen the emergence of new variants of concern, we conducted this study in Bangladesh. The viral RNA genomes extracted from 25 ran- domly collected samples of COVID-19-positive patients from March 2021 to February 2022 were sequenced using Illumina COVID Seq protocol and genomic data processing, as well as evaluations performed in DRAGEN COVID Lineage software. In this study, the percentage of Delta, Omicron, and Mauritius variants identified were 88%, 8%, and 4%, respectively. All of the 25 samples had 23,403 A>G (D614G, S gene), 3037 C>T (nsp3), and 14,408 C>T (nsp12) mutations, where 23,403 A>G was responsible for increased transmis- sion. Omicron had the highest number of unique mutations in the spike protein (i.e., sub- stitutions, deletions, and insertions), which may explain its higher transmissibility and immune-evading ability than Delta. A total of 779 mutations were identified, where 691 substitutions, 85 deletions, and 3 insertion mutations were observed. To sum up, our study will enrich the genomic database of SARS-CoV-2, aiding in treatment strategies along with understanding the virus's preferences in both mutation type and mutation site for predicting newly emerged viruses' survival strategies and thus for preparing to coun- teract them.
Keywords: Bangladesh; COVID-19; Delta; Mauritius; Omicron; SARS-CoV-2; mutation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Evaluation of the Mutational Preferences Throughout the Whole Genome of the Identified Variants of the SARS-CoV-2 Virus Isolates in Bangladesh.Int J Mol Sci. 2025 Jun 25;26(13):6118. doi: 10.3390/ijms26136118. Int J Mol Sci. 2025. PMID: 40649896 Free PMC article.
-
Comprehensive Analysis of SARS-CoV-2 Dynamics in Bangladesh: Infection Trends and Variants (2020-2023).Viruses. 2024 Aug 7;16(8):1263. doi: 10.3390/v16081263. Viruses. 2024. PMID: 39205237 Free PMC article.
-
Molecular Epidemiology of SARS-CoV-2 in Bangladesh.Viruses. 2025 Apr 1;17(4):517. doi: 10.3390/v17040517. Viruses. 2025. PMID: 40284960 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
Cited by
-
Evaluation of the Mutational Preferences Throughout the Whole Genome of the Identified Variants of the SARS-CoV-2 Virus Isolates in Bangladesh.Int J Mol Sci. 2025 Jun 25;26(13):6118. doi: 10.3390/ijms26136118. Int J Mol Sci. 2025. PMID: 40649896 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- figshare/10.6084/m9.figshare.27263484
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

