DNA-Binding Activities of KSHV DNA Polymerase Processivity Factor (PF-8) Complexes
- PMID: 40006945
- PMCID: PMC11860742
- DOI: 10.3390/v17020190
DNA-Binding Activities of KSHV DNA Polymerase Processivity Factor (PF-8) Complexes
Abstract
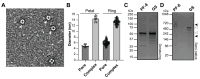
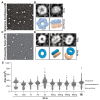
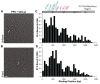
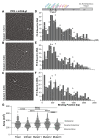
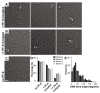
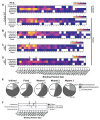
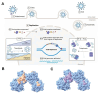
Kaposi's Sarcoma Herpesvirus (KSHV) is the causative agent of several human diseases. There are few effective treatments available to treat infection and KSHV oncogenesis. Disrupting the KSHV infectious cycle would diminish the viral spread. The KSHV lytic phase and production of new virions require efficient copying and packaging of the KSHV genome. KSHV encodes its own lytic DNA replication machinery, including the processivity factor (PF-8), which presents itself as an attractive target for antiviral development. We characterized PF-8 at the single molecule level using transmission electron microscopy to identify key molecular interactions that mediate viral DNA replication initiation. Our results indicate that PF-8 forms oligomeric ring structures (tetramer, hexamer, and/or dodecamer) similar to the related Epstein-Barr virus processivity factor (BMRF1). Our DNA positional mapping revealed high-frequency binding locations of PF-8 within the lytic origin of replication (OriLyt). A multi-variable analysis of PF-8 DNA-binding activity with three mutant OriLyts provides new insights into the mechanisms that PF-8 associates with viral DNA and complexes to form multi-ring-like structures. Collectively, these data enhance the mechanistic understanding of the molecular interactions (protein-protein and protein-DNA) of an essential KSHV DNA replication protein.
Keywords: HHV-8; KSHV; electron microscopy; human herpesviruses; viral replication.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Glycolysis, Glutaminolysis, and Fatty Acid Synthesis Are Required for Distinct Stages of Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication.J Virol. 2017 Apr 28;91(10):e02237-16. doi: 10.1128/JVI.02237-16. Print 2017 May 15. J Virol. 2017. PMID: 28275189 Free PMC article.
-
Inhibiting KSHV replication by targeting the essential activities of KSHV processivity protein, PF-8.J Med Virol. 2024 Oct;96(10):e29958. doi: 10.1002/jmv.29958. J Med Virol. 2024. PMID: 39370884 Free PMC article. Review.
-
FoxO1 Suppresses Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication and Controls Viral Latency.J Virol. 2019 Jan 17;93(3):e01681-18. doi: 10.1128/JVI.01681-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30404794 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Immediate Early Proteins Trigger FOXQ1 Expression in Oral Epithelial Cells, Engaging in a Novel Lytic Cycle-Sustaining Positive Feedback Loop.J Virol. 2023 Mar 30;97(3):e0169622. doi: 10.1128/jvi.01696-22. Epub 2023 Feb 23. J Virol. 2023. PMID: 36815831 Free PMC article.
-
Unraveling the Kaposi Sarcoma-Associated Herpesvirus (KSHV) Lifecycle: An Overview of Latency, Lytic Replication, and KSHV-Associated Diseases.Viruses. 2025 Jan 26;17(2):177. doi: 10.3390/v17020177. Viruses. 2025. PMID: 40006930 Free PMC article. Review.
References
-
- Van Lint A.L., Knipe D.M. Herpesviruses. Encycl. Microbiol. 2019:565–579. doi: 10.1016/B978-0-12-801238-3.02509-5. - DOI
-
- Li S., Bai L., Dong J., Sun R., Lan K. Kaposi’s Sarcoma-Associated Herpesvirus: Epidemiology and Molecular Biology. In: Cai Q., Yuan Z., Lan K., editors. Infectious Agents Associated Cancers: Epidemiology and Molecular Biology Advances in Experimental Medicine and Biology. Vol. 1018. Springer; Singapore: 2017. pp. 91–127. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

