Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Variants in Guinea: A Meta-Analysis of Sequence Data (2020-2023)
- PMID: 40006959
- PMCID: PMC11860642
- DOI: 10.3390/v17020204
Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Variants in Guinea: A Meta-Analysis of Sequence Data (2020-2023)
Abstract
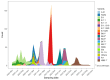
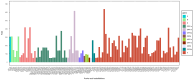
In Guinea, genomic surveillance has been established to generate sequences of and to identify locally circulating SARS-CoV-2 variants. This study aims to describe the distributions, genetic diversity, and origins of SARS-CoV-2 lineages circulating in Guinea during the COVID-19 pandemic. A migration analysis was performed by selecting all sequences generated in Guinea for variants of concern and interest. From March 2020 to December 2023, 1038 sequences were generated in Guinea and submitted to the Global Initiative on Sharing All Influenza Data (GISAID) database. Of these, 73.1% corresponded to SARS-CoV-2 variants of concern, which were further grouped into Omicron (69.4%), Delta (21.9%), Alpha (6.6%), and Eta (2.1%). Other variants accounted for 26.9% of the total. Among the total variants analyzed, 75 importations into Guinea from various countries worldwide were identified. Most of the importations (40%) originated from African countries, followed in significance by those from European countries (25.3%) and Asia (18.6%). A significant migratory flow was observed within Guinea. The genomic surveillance reported in this study revealed the diversity of SARS-CoV-2 variants circulating in Guinea, emphasizing the importance of large-scale sequencing analyses in understanding the dynamics of the pandemic.
Keywords: SARS-CoV-2; genetic diversity; guinea; phylodynamic.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures





Similar articles
-
Nanopore sequencing reveals the genomic diversity of the variants of concern of SARS-CoV-2 during 2021 disease outbreak in Pakistan.Sci Rep. 2025 Aug 1;15(1):28129. doi: 10.1038/s41598-025-12774-1. Sci Rep. 2025. PMID: 40750817 Free PMC article.
-
Genomic epidemiology of SARS-CoV-2 in Norfolk, UK, March 2020-December 2022.Microb Genom. 2025 Jul;11(7):001435. doi: 10.1099/mgen.0.001435. Microb Genom. 2025. PMID: 40663468 Free PMC article.
-
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya.mSphere. 2025 Jun 25;10(6):e0002725. doi: 10.1128/msphere.00027-25. Epub 2025 May 29. mSphere. 2025. PMID: 40439429 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
Cited by
-
The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications.Viruses. 2025 Jul 15;17(7):985. doi: 10.3390/v17070985. Viruses. 2025. PMID: 40733602 Free PMC article. Review.
References
-
- Colson P., Fournier P.-E., Chaudet H., Delerce J., Giraud-Gatineau A., Houhamdi L., Andrieu C., Brechard L., Bedotto M., Prudent E., et al. Analysis of SARS-CoV-2 Variants From 24,181 Patients Exemplifies the Role of Globalization and Zoonosis in Pandemics. Front. Microbiol. 2021;12:786233. doi: 10.3389/fmicb.2021.786233. - DOI - PMC - PubMed
-
- World Health Organization Tracking SARS-CoV-2 Variants. [(accessed on 16 September 2024)]. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants.
-
- Wilkinson E., Giovanetti M., Tegally H., San J.E., Lessells R., Cuadros D., Martin D.P., Rasmussen D.A., Zekri A.-R.N., Sangare A.K., et al. A year of genomic surveillance reveals how the SARS-CoV-2 pandemic unfolded in Africa. Science. 2021;374:423–431. doi: 10.1126/science.abj4336. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

