Targeted Long-Read Sequencing as a Single Assay Improves the Diagnosis of Spastic-Ataxia Disorders
- PMID: 40007153
- PMCID: PMC12040508
- DOI: 10.1002/acn3.70008
Targeted Long-Read Sequencing as a Single Assay Improves the Diagnosis of Spastic-Ataxia Disorders
Abstract
Objective: The hereditary spastic-ataxia spectrum disorders are a group of disabling neurological diseases. The traditional genetic testing pathway is complex, multistep and leaves many cases unsolved. We aim to streamline and improve this process using long-read sequencing.
Methods: We developed a targeted long-read sequencing strategy with the capacity to characterise the genetic variation of all types and sizes within 469 disease-associated genes, in a single assay. We applied this to a cohort of 34 individuals with unsolved spastic-ataxia. An additional five individuals with a known genetic diagnosis were included as positive controls.
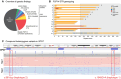
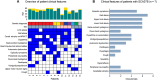
Results: We identified causative pathogenic variants that would be sufficient for genetic diagnosis in 14/34 (41%) unsolved participants. The success rate was 5/11 (45%) in those who were naïve to genetic testing and 9/23 (39%) in those who were undiagnosed after prior genetic testing, completed on a clinical basis. Short tandem repeat expansions in FGF14 were the most common (7/34, 21%). Two individuals (2/34, 6%) had biallelic pathogenic expansions in RFC1 and one individual had a monoallelic pathogenic expansion in ATXN8OS/ATXN8. Causative pathogenic sequence variants other than short tandem repeat expansions were found in four individuals, including in VCP, STUB1, ANO10 and SPG7. Furthermore, all five positive controls were identified.
Interpretation: Our results demonstrate the utility of targeted long-read sequencing in the genetic evaluation of patients with spastic-ataxia spectrum disorders, highlighting both the capacity to increase overall diagnostic yield and to streamline the testing pathway by capturing all known genetic causes in a single assay.
Keywords: hereditary cerebellar ataxia; hereditary spastic paraplegia; nanopore sequencing; spinocerebellar ataxia.
© 2025 The Author(s). Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Kang C., Liang C., Ahmad K. E., et al., “High Degree of Genetic Heterogeneity for Hereditary Cerebellar Ataxias in Australia,” Cerebellum 18, no. 1 (2019): 137–146. - PubMed
-
- Bogdanova‐Mihaylova P., Hebert J., Moran S., et al., “Inherited Cerebellar Ataxias: 5‐Year Experience of the Irish National Ataxia Clinic,” Cerebellum 20, no. 1 (2021): 54–61. - PubMed
-
- Balakrishnan S., Aggarwal S., Muthulakshmi M., et al., “Clinical and Molecular Spectrum of Degenerative Cerebellar Ataxia: A Single Centre Study,” Neurology India 70, no. 3 (2022): 934–942. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
