Large-scale screening of HIV-1 T-cell epitopes restricted by 12 prevalent HLA-A allotypes in Northeast Asia and universal detection of HIV-1-specific CD8+ T cells
- PMID: 40008047
- PMCID: PMC11850406
- DOI: 10.3389/fmicb.2025.1529721
Large-scale screening of HIV-1 T-cell epitopes restricted by 12 prevalent HLA-A allotypes in Northeast Asia and universal detection of HIV-1-specific CD8+ T cells
Abstract
Background: Although the immune response of host T cells to human immunodeficiency virus (HIV) significantly influences the progression of the infection, the development of T-cell-based vaccines and therapies, as well as the clinical evaluation of specific T-cell functions, is currently markedly hindered by the absence of broad-spectrum, functionally validated HIV T-cell epitopes that account for the polymorphisms of the human leukocyte antigen (HLA) within an indicated geographic population. This study aimed to identify T-cell epitopes derived from the GP160, GAG, and POL proteins of the HIV-1 strain, specifically linked to 12 prevalent HLA-A allotypes, that collectively represent approximately 91% of the total gene frequency in Northeast Asian populations.
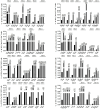
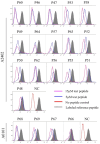
Methods: A total of 134 epitopes were predicted in silico and selected as potential candidates for further validation. Subsequently, peripheral blood mononuclear cells (PBMCs) were collected from 96 individuals with HIV-1 and cocultured ex vivo with each epitope candidate peptide, followed by the detection of activated CD8+ T cells. Peripheral blood mononuclear cells (PBMCs) were collected from 96 individuals with HIV-1 and cocultured ex vivo with each candidate peptide epitope, followed by the detection of activated CD8+ T cells. A total of 69 epitopes were validated as real-world HIV T-cell epitopes presented by 12 dominant HLA-A allotypes. Furthermore, the HLA-A cross-restriction for each epitope candidate was identified through peptide competitive binding assays using 12 transfected HMy2.CIR cell lines.
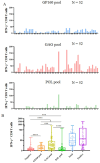
Results: A total of 45 epitopes demonstrated high affinity, while 31 epitopes displayed intermediate affinity. A broad-spectrum CD8+ T-cell epitope library containing 141 validated epitope peptides was used to universally detect HIV-1-specific CD8+ T cells via peptide-PBMC ex vivo coculture and intracellular IFN-γ staining. In 52 people with HIV-1, the number of reactive HIV-1 specific CD8+ T cells was significantly higher in the CD4+ T-cell-high patient group compared to the CD4+ T-cell-low patient group, and it correlated with the CD4+ T-cell-low patient group (<200/μL).
Conclusion: This study provides a broad-spectrum CD8+ T-cell epitope library aimed at developing a T-cell-directed HIV vaccine that offers high population coverage in Northeast Asia. In addition, it establishes a universal detection method for the clinical assessment of HIV-1-specific CD8+ T-cell responses.
Keywords: CD8+ T-cell; HIV-1; HLA-A; T-cell epitope; antigen-specific T-cell detection.
Copyright © 2025 Ding, Yan, Huang, Yu, Wu, Shen and Fang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Screening and Identification of HBV Epitopes Restricted by Multiple Prevalent HLA-A Allotypes.Front Immunol. 2022 Apr 7;13:847105. doi: 10.3389/fimmu.2022.847105. eCollection 2022. Front Immunol. 2022. PMID: 35464415 Free PMC article.
-
Predominant T-cell epitopes of SARS-CoV-2 restricted by multiple prevalent HLA-B and HLA-C allotypes in Northeast Asia.Front Immunol. 2025 May 21;16:1545510. doi: 10.3389/fimmu.2025.1545510. eCollection 2025. Front Immunol. 2025. PMID: 40469303 Free PMC article.
-
Large-Scale Screening of CD4+ T-Cell Epitopes From SARS-CoV-2 Proteins and the Universal Detection of SARS-CoV-2 Specific T Cells for Northeast Asian Population.J Med Virol. 2025 Feb;97(2):e70241. doi: 10.1002/jmv.70241. J Med Virol. 2025. PMID: 39977358
-
Identification and selection of immunodominant B and T cell epitopes for dengue multi-epitope-based vaccine.Med Microbiol Immunol. 2021 Feb;210(1):1-11. doi: 10.1007/s00430-021-00700-x. Epub 2021 Jan 30. Med Microbiol Immunol. 2021. PMID: 33515283 Review.
-
Multiple Approaches for Increasing the Immunogenicity of an Epitope-Based Anti-HIV Vaccine.AIDS Res Hum Retroviruses. 2015 Nov;31(11):1077-88. doi: 10.1089/AID.2015.0101. Epub 2015 Aug 13. AIDS Res Hum Retroviruses. 2015. PMID: 26149745 Review.
Cited by
-
A systematic review of T cell epitopes defined from the proteome of human immunodeficiency virus.Virus Res. 2025 Aug;358:199602. doi: 10.1016/j.virusres.2025.199602. Epub 2025 Jun 23. Virus Res. 2025. PMID: 40562176 Free PMC article. Review.
References
-
- Addo M. M., Yu X. G., Rathod A., Cohen D., Eldridge R. L., Strick D., et al. . (2003). Comprehensive epitope analysis of human immunodeficiency virus type 1 (hiv-1)-specific t-cell responses directed against the entire expressed hiv-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77, 2081–2092. doi: 10.1128/jvi.77.3.2081-2092.2003, PMID: - DOI - PMC - PubMed
-
- Bailón L., Llano A., Cedeño S., Escribà T., Rosás-Umbert M., Parera M., et al. . (2022). Safety, immunogenicity and effect on viral rebound of hti vaccines in early treated hiv-1 infection: a randomized, placebo-controlled phase 1 trial. Nat. Med. 28, 2611–2621. doi: 10.1038/s41591-022-02060-2, PMID: - DOI - PubMed
-
- Cheng Y., Gunasegaran B., Singh H. D., Dutertre C. A., Loh C. Y., Lim J. Q., et al. . (2021). Non-terminally exhausted tumor-resident memory hbv-specific t cell responses correlate with relapse-free survival in hepatocellular carcinoma. Immunity 54, 1825–1840.e7. doi: 10.1016/j.immuni.2021.06.013, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

