Genome-Wide Association Study for Resting Electrocardiogram in the Qatari Population Identifies 6 Novel Genes and Validates Novel Polygenic Risk Scores
- PMID: 40008532
- PMCID: PMC12132764
- DOI: 10.1161/JAHA.124.038341
Genome-Wide Association Study for Resting Electrocardiogram in the Qatari Population Identifies 6 Novel Genes and Validates Novel Polygenic Risk Scores
Abstract
Background: Electrocardiography is one of the most valuable noninvasive diagnostic tools in determining the presence of many cardiovascular diseases. Genetic factors are important in determining ECG abnormalities and their link to cardiovascular diseases. Genome-wide association studies and polygenic risk scores (PRSs) have been conducted for various ECG traits such as QT interval and QRS duration. However, these studies mainly focused on cohorts of European descent.
Methods: In this cohort study, genome-wide association studies for 6 ECG traits (RR, PR, corrected QT interval [QTc], QRS, JT, and P wave duration) were conducted in a Middle Eastern cohort from the Qatar Precision Health Institute, comprising 13 827 subjects with whole-genome sequence data. Middle Eastern PRSs were developed using clumping and thresholding, and their performance was compared with 26 published PRSs. Genetic predisposition to long QT syndrome was explored using rare variant analysis.
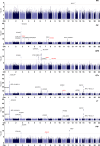
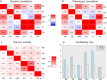
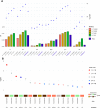
Results: Seventy-four independent loci were obtained with genome-wide significance across the 6 traits (P<5×10-8). Of the 74 loci, 67 (90.5%) were previously reported, and 7 loci (9.5%) were novel and contained 6 genes: STAC and CSMD1 for PR, ANK1 and NCOA2 for QRS, LSP1 for QTc, and MKLN1 for P wave duration. All 26 published PRSs showed good performance in our cohort. PGS002276 showed the best performance for QTc (R2=0.059, P=4.83×10-185), PGS002166 showed the best performance for QRS (R2=0.024, P=1.53×10-75), and PGS000905 showed the best performance for PR (R2=0.053, P=2.57×10-165). Some of these PRSs were associated with cardiovascular diseases. For example, PGS003500, a QTc PRS, was significantly associated with cardiomyopathy (odds ratio per 1 SD=1.58 [95% CI, 1.23-2.01]; P=2.42×10-4). Middle Eastern PRSs substantially outperformed published PRSs and did not perform well in the UK Biobank data. Ten pathogenic variants, including 3 that are specific to Qatari individuals, were observed in 17 long QT syndrome genes and were carried by 19 individuals. The QTc average was larger for mutation carriers (415.6±23.5 versus 402.3±18.5 in noncarriers). Five-year follow-up data did not show a significant change in ECG patterns, regardless of mutation status and PRS values. Four of 2302 individuals had prolonged QTc intervals over the 2 time points.
Conclusions: In this first genome-wide association study for ECG traits in the Middle East using whole-genome sequence data, 7 novel loci (6 genes) were identified. Published PRSs performed well, but newly developed Middle Eastern-specific PRSs performed the best. Novel variants in long QT syndrome genes were observed for the first time in Qatari individuals. Follow-up data did not show significant changes in ECG patterns.
Keywords: Middle East; Qatar precision health institute; cardiovascular diseases; diverse populations; electrocardiography; genome‐wide association studies; polygenic risk scores.
Conflict of interest statement
None.
Figures


Similar articles
-
Monogenic and Polygenic Contributions to QTc Prolongation in the Population.Circulation. 2022 May 17;145(20):1524-1533. doi: 10.1161/CIRCULATIONAHA.121.057261. Epub 2022 Apr 7. Circulation. 2022. PMID: 35389749 Free PMC article.
-
Transethnic Genome-Wide Association Study Provides Insights in the Genetic Architecture and Heritability of Long QT Syndrome.Circulation. 2020 Jul 28;142(4):324-338. doi: 10.1161/CIRCULATIONAHA.120.045956. Epub 2020 May 20. Circulation. 2020. PMID: 32429735 Free PMC article.
-
Polygenic risk scores for the prediction of common cancers in East Asians: A population-based prospective cohort study.Elife. 2023 Mar 27;12:e82608. doi: 10.7554/eLife.82608. Elife. 2023. PMID: 36971353 Free PMC article.
-
Precision Medicine in Cardiovascular Disease Prevention: Clinical Validation of Multi-Ancestry Polygenic Risk Scores in a U.S. Cohort.Nutrients. 2025 Mar 6;17(5):926. doi: 10.3390/nu17050926. Nutrients. 2025. PMID: 40077796 Free PMC article.
-
Polygenic risk scores in kidney transplantation.Curr Opin Organ Transplant. 2025 Jun 1;30(3):208-214. doi: 10.1097/MOT.0000000000001212. Epub 2025 Apr 1. Curr Opin Organ Transplant. 2025. PMID: 40171629 Review.
References
-
- Ntalla I, Weng LC, Cartwright JH, Hall AW, Sveinbjornsson G, Tucker NR, Choi SH, Chaffin MD, Roselli C, Barnes MR, et al. Multi‐ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction. Nat Commun. 2020;11:2542. doi: 10.1038/s41467-020-15706-x - DOI - PMC - PubMed
-
- Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–836. doi: 10.1038/ng.3014 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

