Inhibition of pneumococcal growth and biofilm formation by human isolates of Streptococcus mitis and Streptococcus oralis
- PMID: 40008876
- PMCID: PMC11921387
- DOI: 10.1128/aem.01336-24
Inhibition of pneumococcal growth and biofilm formation by human isolates of Streptococcus mitis and Streptococcus oralis
Abstract
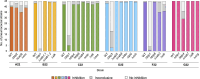
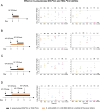
In a world facing the unprecedented threat of antibiotic-resistant bacteria, targeted approaches to control colonization and prevent disease caused by common pathobionts offer a promising solution. Streptococcus pneumoniae (pneumococcus) is a leading cause of infections worldwide, affecting both children and adults despite available antimicrobials and vaccines. Colonization, which occurs in the form of a biofilm in the upper respiratory tract, is frequent and a prerequisite for disease and transmission. The use of live bacterial strains as biotherapeutics for infectious diseases is actively being explored. Here, we investigated the potential of commensal streptococci to control S. pneumoniae. Screening of over 300 human isolates led to the identification of seven strains (one Streptococcus oralis and six Streptococcus mitis, designated A22 to G22) with inhibitory activity against S. pneumoniae of multiple serotypes and genotypes. Characterization of A22 to G22 cell-free supernatants indicated the involvement of secreted proteins or peptides in the inhibitory effect of all S. mitis isolates. Genome analyses revealed the presence of 64 bacteriocin loci, encoding 70 putative bacteriocins, several of which are novel and absent or rare in over 7,000 publicly available pneumococcal genomes. Deletion mutants indicated that bacteriocins partially or completely explained the anti-pneumococcal activity of the commensal strains. Importantly, strains A22 to G22 were further able to prevent and disrupt pneumococcal biofilms, a proxy for nasopharyngeal colonization. These results highlight the intricacy of the interactions among nasopharyngeal colonizers and support the potential of strains A22 to G22 to be used as live biotherapeutics, alone or in combination, to control S. pneumoniae colonization.
Importance: Streptococcus pneumoniae (pneumococcus) infections remain a major public health issue despite the use of vaccines and antibiotics. Pneumococci asymptomatically colonize the human upper respiratory tract, a niche shared with several commensal Streptococcus species. Competition for space and nutrients among species sharing the same niche is well documented and tends to be more intense among closely related species. Based on this rationale, a screening of several commensal streptococci isolated from the human upper respiratory tract led to the identification of strains of Streptococcus mitis and Streptococcus oralis capable of inhibiting most pneumococcal strains, across diverse serotypes and genotypes. This inhibition was partially or wholly linked to the expression of novel bacteriocins. The selected S. mitis and S. oralis strains significantly disrupted pneumococcal biofilms, indicating a potential for using commensals as biotherapeutics to control pneumococcal colonization, a key step in preventing disease and transmission.
Keywords: Streptococcus mitis; Streptococcus oralis; Streptococcus pneumoniae; bacteriocin; biofilm; biotherapeutic; colonization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Streptococcus pneumoniae serotypes that frequently colonise the human nasopharynx are common recipients of penicillin-binding protein gene fragments from Streptococcus mitis.Microb Genom. 2021 Sep;7(9):000622. doi: 10.1099/mgen.0.000622. Microb Genom. 2021. PMID: 34550067 Free PMC article.
-
Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of "Atypical" pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes.Infect Immun. 2000 Mar;68(3):1374-82. doi: 10.1128/IAI.68.3.1374-1382.2000. Infect Immun. 2000. PMID: 10678950 Free PMC article.
-
Identification of Virulence-Associated Properties by Comparative Genome Analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, Three S. oralis Subspecies, and S. infantis.mBio. 2019 Sep 3;10(5):e01985-19. doi: 10.1128/mBio.01985-19. mBio. 2019. PMID: 31481387 Free PMC article.
-
Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease.Front Cell Infect Microbiol. 2015 Jan 13;4:194. doi: 10.3389/fcimb.2014.00194. eCollection 2014. Front Cell Infect Microbiol. 2015. PMID: 25629011 Free PMC article. Review.
-
Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci.FEMS Microbiol Rev. 2009 May;33(3):572-86. doi: 10.1111/j.1574-6976.2009.00172.x. FEMS Microbiol Rev. 2009. PMID: 19396958 Review.
Cited by
-
Genomic and functional insights into commensal streptococci with anti-pneumococcal activity.BMC Genomics. 2025 Jul 1;26(1):577. doi: 10.1186/s12864-025-11756-x. BMC Genomics. 2025. PMID: 40596837 Free PMC article.
-
Bioinformatics-driven discovery of skin microbiota bacteriocins as potential antibiotics and probiotics.J Antibiot (Tokyo). 2025 Jul 24. doi: 10.1038/s41429-025-00847-2. Online ahead of print. J Antibiot (Tokyo). 2025. PMID: 40702306
-
Molecular biology insights into levofloxacin-loaded ZnO nanoparticles: a potent strategy against MDR Acinetobacter baumannii.RSC Adv. 2025 Aug 26;15(37):30189-30201. doi: 10.1039/d5ra03081a. eCollection 2025 Aug 22. RSC Adv. 2025. PMID: 40874159 Free PMC article.
References
-
- Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, Lukšić I, Nair H, McAllister DA, Campbell H, Rudan I, Black R, Knoll MD. 2018. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health 6:e744–e757. doi:10.1016/S2214-109X(18)30247-X - DOI - PMC - PubMed
-
- Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, Gladstone RA, Turner P, Keenan JD, Breiman RF, Nahm MH. 2020. A new pneumococcal capsule type, 10D, is the 100th Serotype and has a large cps fragment from an oral streptococcus. MBio 11:e00937-20. doi:10.1128/mBio.00937-20 - DOI - PMC - PubMed
-
- Ganaie F, Maruhn K, Li C, Porambo RJ, Elverdal PL, Abeygunwardana C, van der Linden M, Duus JØ, Sheppard CL, Nahm MH. 2021. Structural, genetic, and serological elucidation of Streptococcus pneumoniae serogroup 24 serotypes: discovery of a new serotype, 24C, with a variable capsule structure. J Clin Microbiol 59:e0054021. doi:10.1128/JCM.00540-21 - DOI - PMC - PubMed
-
- Essink B, Sabharwal C, Cannon K, Frenck R, Lal H, Xu X, Sundaraiyer V, Peng Y, Moyer L, Pride MW, Scully IL, Jansen KU, Gruber WC, Scott DA, Watson W. 2022. Pivotal phase 3 randomized clinical trial of the safety, tolerability, and immunogenicity of 20-valent pneumococcal conjugate vaccine in adults aged ≥18 Years. Clin Infect Dis 75:390–398. doi:10.1093/cid/ciab990 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- PTDC/BIA-MIC/30703/2017/Fundacao para a Ciencia e a Tecnologia
- UIDB/04612/2020/Fundacao para a Ciencia e a Tecnologia
- LA/P/0097/2020/Fundacao para a Ciencia e a Tecnologia
- 2021.07866.BD PD/BD/128365/2017 UI/BD/153385/20222 2020.05293.BD PD/BD/148434/2019 SFRH/BPD/115280/2/Fundacao para a Ciencia e a Tecnologia
LinkOut - more resources
Full Text Sources

