Identification of immunogenic HLA-A*02:01 epitopes associated with HCC for immunotherapy development
- PMID: 40008881
- PMCID: PMC11868434
- DOI: 10.1097/HC9.0000000000000659
Identification of immunogenic HLA-A*02:01 epitopes associated with HCC for immunotherapy development
Abstract
Background: HCC is the most common form of primary liver cancer, and despite recent advances in cancer treatment, it remains associated with poor prognosis and a lack of response to conventional therapies. Immunotherapies have emerged as a promising approach for cancer treatment, especially through the identification of tumor-specific immunogenic epitopes that can trigger a targeted immune response. This study aimed to identify immunogenic epitopes associated with HCC for the development of specific immunotherapies.
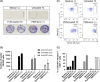
Methods: We used high-throughput data screening and bioinformatics tools for antigens and epitope selection. The immunogenicity of the selected epitopes was studied after coculture of peripheral blood mononuclear cells obtained from healthy donors or HCC patients with a plasmacytoid dendritic cell line loaded with the selected peptides. Specific CD8+ T cell amplification and functionality were determined by labeling with tetramers and by IFN-γ and CD107a expression (flow cytometry and ELISpot).
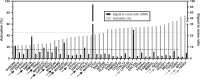
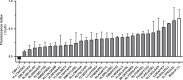
Results: We analyzed the transcriptional gene expression landscape of HCC to screen for a set of 16 ectopically expressed genes in a majority of HCC samples. Epitopes predicted to bind to HLA-A*02:01 with high affinity were further validated for their immunogenicity using the previously described plasmacytoid dendritic cell line in ex vivo CD8+ activation assays using patient immune cells. Three out of the 30 tested epitopes, namely FLWGPRALV (MAGE-A3), FMNKFIYEI (AFP), and KMFHTLDEL (LRRC46), elicited a strong T-cell response, in activation assays, degranulation assays, and IFN-γ secretion assays.
Conclusions: These results highlight the potential of these peptides to be considered as targets for immunotherapies. The discovery of such immunogenic epitopes should improve immune-based treatments for liver cancer in combination with the current treatment approach.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Dr Joel Plumas is a cofounder, shareholder, and employee of PDC*line Pharma. Thomas Decaens consults, advises, and receives grants from BMS, AstraZeneca, and Roche. He consults and receives grants from Gilead and Abbvie. He received grants from Geoscience and Netris Pharma. He consults for BD. The remaining authors have no conflicts to report.
Figures


References
-
- Qin S, Chen M, Cheng AL, Kaseb AO, Kudo M, Lee HC, et al. . Atezolizumab plus bevacizumab versus active surveillance in patients with resected or ablated high-risk hepatocellular carcinoma (IMbrave050): A randomised, open-label, multicentre, phase 3 trial. Lancet. 2023;402:1835–1847. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

