Integrative single-cell and spatial transcriptome analysis reveals heterogeneity of human liver progenitor cells
- PMID: 40008906
- PMCID: PMC11868439
- DOI: 10.1097/HC9.0000000000000662
Integrative single-cell and spatial transcriptome analysis reveals heterogeneity of human liver progenitor cells
Abstract
Background: Liver progenitor cells (LPCs) with bipotential differentiation capacities are essential for restoring liver homeostasis and hepatocyte population after damage. However, the low proportion and shared markers with epithelial cells make studying LPC heterogeneity difficult, especially in humans. To address this gap, we explored over 259,400 human liver single cells across 4 conditions (fetal, healthy, cirrhotic, and HCC-affected livers).
Methods: Human liver tissue samples were analyzed using spatial transcriptomics sequencing technologies to describe the heterogeneity of LPCs. Liver tissue was characterized by LPC heterogeneity at single-cell resolution by employing cellular modules, differentiation trajectories, and gene co-expression patterns.
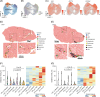
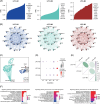
Results: We annotated and identified 1 LPC cluster, 3 LPC subpopulations, and 4 distinct cellular modules, indicating the heterogeneity within LPC and the diversity between LPCs and epithelial cells. LPCs showed spatial colocalization with cholangiocytes and comprised a small proportion (2.95±1.91%) within the merged epithelial cells and LPC populations, exhibiting marked differences in marker expression patterns compared to those in mice. LPCs exhibited distinct cellular states in functional restoration, activation, proliferation, and cell transition. Additionally, the gene co-expression network of LPCs exhibited 3 unique modules, reflecting the distinct connectivity of genes encoding apolipoproteins and heat shock proteins in the gene co-expression network modules.
Conclusions: Our study provides valuable insights into the multifaceted heterogeneity of human LPCs. Future studies focusing on spatial gene expression dynamics will contribute to our understanding of the spatial arrangement of liver regeneration.
Copyright © 2025 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Bhaskar Roy received grants from the National Key R&D program. The remaining authors have no conflicts to report.
Figures




Similar articles
-
Hdac1 Regulates Differentiation of Bipotent Liver Progenitor Cells During Regeneration via Sox9b and Cdk8.Gastroenterology. 2019 Jan;156(1):187-202.e14. doi: 10.1053/j.gastro.2018.09.039. Epub 2018 Sep 26. Gastroenterology. 2019. PMID: 30267710 Free PMC article.
-
Targeted deletion of ER chaperone GRP94 in the liver results in injury, repopulation of GRP94-positive hepatocytes, and spontaneous hepatocellular carcinoma development in aged mice.Neoplasia. 2014 Aug;16(8):617-26. doi: 10.1016/j.neo.2014.07.005. Neoplasia. 2014. PMID: 25220589 Free PMC article.
-
Identification of a Liver Progenitor Cell-Related Genes Signature Predicting Overall Survival for Hepatocellular Carcinoma.Technol Cancer Res Treat. 2021 Jan-Dec;20:15330338211041425. doi: 10.1177/15330338211041425. Technol Cancer Res Treat. 2021. PMID: 34866477 Free PMC article.
-
Liver progenitor cell-driven liver regeneration.Exp Mol Med. 2020 Aug;52(8):1230-1238. doi: 10.1038/s12276-020-0483-0. Epub 2020 Aug 14. Exp Mol Med. 2020. PMID: 32796957 Free PMC article. Review.
-
The progenitor cell dilemma: Cellular and functional heterogeneity in assistance or escalation of liver injury.J Hepatol. 2017 Mar;66(3):619-630. doi: 10.1016/j.jhep.2016.10.033. Epub 2016 Nov 5. J Hepatol. 2017. PMID: 27826058 Review.
References
-
- Lukacs-Kornek V, Lammert F. The progenitor cell dilemma: Cellular and functional heterogeneity in assistance or escalation of liver injury. J Hepatol. 2017;66:619–630. - PubMed
-
- Sharma A, Seow JJW, Dutertre CA, Pai R, Blériot C, Mishra A, et al. . Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

