A tumor-binding antibody with cross-reactivity to viral antigens
- PMID: 40009215
- PMCID: PMC11865367
- DOI: 10.1007/s00262-025-03975-8
A tumor-binding antibody with cross-reactivity to viral antigens
Abstract
Background: We previously identified in non-small cell lung cancer (NSCLC) patients an autoantibody to complement factor H (CFH) that is associated with non-metastatic disease and longer time to progression in patients with stage I disease. A recombinant human antibody, GT103, was cloned from single B cells isolated from patients with the autoantibody. GT103 inhibits tumor growth and establishes an antitumor microenvironment. The anti-CFH autoantibody and GT103 recognize the epitope PIDNGDIT within the SCR19 domain of CFH. Here, we asked if this autoantibody could have originally arisen as a humoral response to a similar epitope in a viral protein from a prior infection.
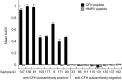
Methods: Homologous viral peptides with high sequence identity to the core PIDNGDIT epitope sequence were identified and synthesized. NSCLC patient plasma containing anti-CFH autoantibodies were assayed by ELISA against these peptides. GT103 was assayed on a 4345-peptide pathogen microarray.
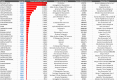
Results: Epitopes similar to the GT103 epitope are present in several viruses, including human metapneumovirus-1 (HMPV-1) that contains a sequence within attachment glycoprotein G that differs by one amino acid. Anti-CFH autoantibodies in NSCLC patient plasma weakly bound to an HMPV-1 peptide containing the epitope. GT103 cross-reacted with multiple viral epitopes on a peptide microarray, with the top hits being peptides in the human endogenous retrovirus-K polymerase (HERV-K pol) protein and measles hemagglutinin glycoprotein. GT103 bound the viral HMPV-1, HERV-K pol, and measles epitope peptides but with lower affinity compared to the GT103 epitope peptide.
Conclusion: These findings suggest that memory B cells against a viral target could have affinity matured to produce an antibody that recognizes a similar epitope on tumor cells and exhibits antitumor properties.
Keywords: Antibody cross-reactivity; Autoantibodies; Humoral immunity; Molecular mimicry; Viral epitopes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: E.F.P., E.B.G., and M.J.C. are founders of Grid Therapeutics. E.F.P. is the CEO of Grid Therapeutics. K.W. has no competing interests to declare that are relevant to the content of this article.
Figures

References
-
- Wang D, Yang D, Yang L et al (2023) Human autoantigen atlas: searching for the hallmarks of autoantigens. J Proteome Res 22:1800–1815. 10.1021/acs.jproteome.2c00799 - PubMed
-
- Yaniv G, Twig G, Shor DB et al (2015) A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients. Autoimmun Rev 14:75–79. 10.1016/j.autrev.2014.10.003 - PubMed
-
- Malmstrom V, Catrina AI, Klareskog L (2017) The immunopathogenesis of seropositive rheumatoid arthritis: from triggering to targeting. Nat Rev Immunol 17:60–75. 10.1038/nri.2016.124 - PubMed
-
- Bian X, Wasserfall C, Wallstrom G et al (2017) Tracking the antibody immunome in type 1 diabetes using protein arrays. J Proteome Res 16:195–203. 10.1021/acs.jproteome.6b00354 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

