The synaptonemal complex aligns meiotic chromosomes by wetting
- PMID: 40009663
- PMCID: PMC11864179
- DOI: 10.1126/sciadv.adt5675
The synaptonemal complex aligns meiotic chromosomes by wetting
Abstract
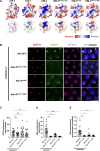
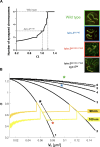
During meiosis, the parental chromosomes are drawn together to enable exchange of genetic information. Chromosomes are aligned through the assembly of a conserved interface, the synaptonemal complex, composed of a central region that forms between two parallel chromosomal backbones called axes. Here, we identify the axis-central region interface in C. elegans, containing a conserved positive patch on the axis component HIM-3 and the negative C terminus of the central region protein SYP-5. Crucially, the canonical ultrastructure of the synaptonemal complex is altered upon weakening this interface using charge-reversal mutations. We developed a thermodynamic model that recapitulates our experimental observations, indicating that the liquid-like central region can assemble by wetting the axes without active energy consumption. More broadly, our data show that condensation drives tightly regulated nuclear reorganization during sexual reproduction.
Figures




Update of
-
The synaptonemal complex aligns meiotic chromosomes by wetting.bioRxiv [Preprint]. 2024 Sep 27:2024.08.07.607092. doi: 10.1101/2024.08.07.607092. bioRxiv. 2024. Update in: Sci Adv. 2025 Feb 28;11(9):eadt5675. doi: 10.1126/sciadv.adt5675. PMID: 39149313 Free PMC article. Updated. Preprint.
References
-
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). - PubMed
-
- Gouveia B., Kim Y., Shaevitz J. W., Petry S., Stone H. A., Brangwynne C. P., Capillary forces generated by biomolecular condensates. Nature 609, 255–264 (2022). - PubMed
-
- Strom A. R., Kim Y., Zhao H., Chang Y.-C., Orlovsky N. D., Košmrlj A., Storm C., Brangwynne C. P., Condensate interfacial forces reposition DNA loci and probe chromatin viscoelasticity. Cell 187, 5282–5297.e20 (2024). - PubMed
-
- Zickler D., Kleckner N., Meiosis: Dances between homologs. Annu. Rev. Genet. 57, 1–63 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

