Complete Blood Count and Monocyte Distribution Width-Based Machine Learning Algorithms for Sepsis Detection: Multicentric Development and External Validation Study
- PMID: 40009841
- PMCID: PMC11904381
- DOI: 10.2196/55492
Complete Blood Count and Monocyte Distribution Width-Based Machine Learning Algorithms for Sepsis Detection: Multicentric Development and External Validation Study
Abstract
Background: Sepsis is an organ dysfunction caused by a dysregulated host response to infection. Early detection is fundamental to improving the patient outcome. Laboratory medicine can play a crucial role by providing biomarkers whose alteration can be detected before the onset of clinical signs and symptoms. In particular, the relevance of monocyte distribution width (MDW) as a sepsis biomarker has emerged in the previous decade. However, despite encouraging results, MDW has poor sensitivity and positive predictive value when compared to other biomarkers.
Objective: This study aims to investigate the use of machine learning (ML) to overcome the limitations mentioned earlier by combining different parameters and therefore improving sepsis detection. However, making ML models function in clinical practice may be problematic, as their performance may suffer when deployed in contexts other than the research environment. In fact, even widely used commercially available models have been demonstrated to generalize poorly in out-of-distribution scenarios.
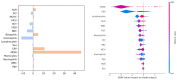
Methods: In this multicentric study, we developed ML models whose intended use is the early detection of sepsis on the basis of MDW and complete blood count parameters. In total, data from 6 patient cohorts (encompassing 5344 patients) collected at 5 different Italian hospitals were used to train and externally validate ML models. The models were trained on a patient cohort encompassing patients enrolled at the emergency department, and it was externally validated on 5 different cohorts encompassing patients enrolled at both the emergency department and the intensive care unit. The cohorts were selected to exhibit a variety of data distribution shifts compared to the training set, including label, covariate, and missing data shifts, enabling a conservative validation of the developed models. To improve generalizability and robustness to different types of distribution shifts, the developed ML models combine traditional methodologies with advanced techniques inspired by controllable artificial intelligence (AI), namely cautious classification, which gives the ML models the ability to abstain from making predictions, and explainable AI, which provides health operators with useful information about the models' functioning.
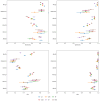
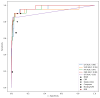
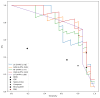
Results: The developed models achieved good performance on the internal validation (area under the receiver operating characteristic curve between 0.91 and 0.98), as well as consistent generalization performance across the external validation datasets (area under the receiver operating characteristic curve between 0.75 and 0.95), outperforming baseline biomarkers and state-of-the-art ML models for sepsis detection. Controllable AI techniques were further able to improve performance and were used to derive an interpretable set of diagnostic rules.
Conclusions: Our findings demonstrate how controllable AI approaches based on complete blood count and MDW may be used for the early detection of sepsis while also demonstrating how the proposed methodology can be used to develop ML models that are more resistant to different types of data distribution shifts.
Keywords: artificial intelligence; biomarker; clinical signs; clinical symptoms; complete blood count; controllable AI; data distribution; detection; development study; diagnostic; early detection; external validation; machine learning; machine learning model; medical machine learning; organ; organ dysfunction; sepsis; sepsis detection; validation study.
©Andrea Campagner, Luisa Agnello, Anna Carobene, Andrea Padoan, Fabio Del Ben, Massimo Locatelli, Mario Plebani, Agostino Ognibene, Maria Lorubbio, Elena De Vecchi, Andrea Cortegiani, Elisa Piva, Donatella Poz, Francesco Curcio, Federico Cabitza, Marcello Ciaccio. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 26.02.2025.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


References
-
- Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016 Feb 23;315(8):801–10. doi: 10.1001/jama.2016.0287. https://europepmc.org/abstract/MED/26903338 2492881 - DOI - PMC - PubMed
-
- Seymour CW, Gesten F, Prescott HC, Friedrich ME, Iwashyna TJ, Phillips GS, Lemeshow S, Osborn T, Terry KM, Levy MM. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 2017 Jun 08;376(23):2235–44. doi: 10.1056/NEJMoa1703058. https://europepmc.org/abstract/MED/28528569 - DOI - PMC - PubMed
-
- Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, Machado FR, Mcintyre L, Ostermann M, Prescott HC, Schorr C, Simpson S, Wiersinga WJ, Alshamsi F, Angus DC, Arabi Y, Azevedo L, Beale R, Beilman G, Belley-Cote E, Burry L, Cecconi M, Centofanti J, Coz Yataco A, De Waele J, Dellinger RP, Doi K, Du B, Estenssoro E, Ferrer R, Gomersall C, Hodgson C, Møller MH, Iwashyna T, Jacob S, Kleinpell R, Klompas M, Koh Y, Kumar A, Kwizera A, Lobo S, Masur H, McGloughlin S, Mehta S, Mehta Y, Mer M, Nunnally M, Oczkowski S, Osborn T, Papathanassoglou E, Perner A, Puskarich M, Roberts J, Schweickert W, Seckel M, Sevransky J, Sprung CL, Welte T, Zimmerman J, Levy M. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021 Nov;47(11):1181–247. doi: 10.1007/s00134-021-06506-y. https://europepmc.org/abstract/MED/34599691 10.1007/s00134-021-06506-y - DOI - PMC - PubMed
-
- UK NSC: evidence review process. United Kingdom Government. 2024. Jul 26, [2025-01-10]. https://www.gov.uk/government/publications/uk-nsc-evidence-review-proces... .
-
- Fan SL, Miller NS, Lee J, Remick DG. Diagnosing sepsis - the role of laboratory medicine. Clin Chim Acta. 2016 Sep 01;460:203–10. doi: 10.1016/j.cca.2016.07.002. https://europepmc.org/abstract/MED/27387712 S0009-8981(16)30293-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

