Speeding up drug susceptibility testing in Mycobacterium tuberculosis using RNA biomarkers
- PMID: 40010155
- PMCID: PMC11905850
- DOI: 10.1016/j.ebiom.2025.105611
Speeding up drug susceptibility testing in Mycobacterium tuberculosis using RNA biomarkers
Abstract
Background: Efficient management of drug-resistant tuberculosis relies on fast diagnostics. To accelerate phenotypic drug susceptibility testing [pDST] for Mycobacterium tuberculosis [TB], we introduce TRACeR-TB, a test that infers drug resistance from antibiotic-specific mRNA biomarkers.
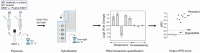
Methods: To develop TRACeR-TB, target genes were first identified through RNA sequencing experiments conducted on two drug-exposed, susceptible strains for four antitubercular drugs. Based on these findings, we designed drug-specific multiplex Quantigene panels to quantify mRNA levels of 8-9 biomarkers per drug (class), directly from crude cell lysates. The performance of TRACeR-TB was compared to the widely used Mycobacteria Growth Indicator Tube [MGIT] pDST by subjecting 238 strains with diverse drug resistance profiles to both methods, and aligning results to genotypic data. Furthermore, we explored TRACeR-TB's potential for evaluating molecules that enhance antibiotic efficacy, and investigated its applicability in macrophage models to assess Mtb's intracellular stress responses to drugs.
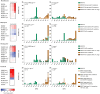
Findings: Antituberculosis drugs trigger distinct transcriptional stress responses in susceptible, but not resistant bacilli, enabling a differentiation of the antibiotic phenotype in only 6 h. Validation on 238 strains showed TRACeR-TB had 100% (95% CI: 93·1-100%) sensitivity and 89·5% (95% CI: 74·7-97·2%) specificity compared to, respectively, 82·3% (95% CI: 69·2%-91·5%) and 94·8% (95% CI: 81·9%-99·4%) for MGIT pDST. TRACeR-TB specificity is likely underestimated due to the inclusion of isolates harbouring uncharacterised mutations. TRACeR-TB demonstrated 100% concordance with MGIT for drugs with reliable MGIT outcomes (moxifloxacin and isoniazid). Additionally, its sensitivity outperformed current rifampicin testing, detecting resistance in all borderline-resistant strains that MGIT missed, and bedaquiline testing. Furthermore, the assay detected the predicted effect of a novel drug booster and the intracellular drug-induced stress in macrophage models, highlighting its potential for drug optimisation.
Interpretation: TRACeR-TB is a complementary addition to current DSTs and can have a substantial impact on the TB diagnostics field. This tool can also play a vital role in identifying resistance mutations, thereby closing gaps in genotypic knowledge, and contribute to drug discovery and development.
Funding: Institut Pasteur, Agence Nationale de la Recherche.
Keywords: Antimicrobial resistance; Biomarkers; Diagnosis; RNA; Tuberculosis.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Authors declare no conflict of interests regarding any financial and personal relationships with other people or organisations that could inappropriately influence our work.
Figures

References
-
- World Health Organization . Global Tuberculosis Report 2023. World Health Organization; Geneva: 2023.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

