Control of plasma membrane-associated actin polymerization specifies the pattern of the cell wall in xylem vessels
- PMID: 40011437
- PMCID: PMC11865516
- DOI: 10.1038/s41467-025-56866-y
Control of plasma membrane-associated actin polymerization specifies the pattern of the cell wall in xylem vessels
Abstract
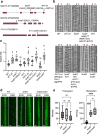
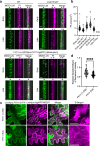
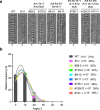
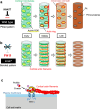
Cell wall patterning is central to determining the shape and function of plant cells. Protoxylem and metaxylem vessel cells deposit banded and pitted cell walls, respectively, which enable their distinctive water transport capabilities. Here, we show that the pitted cell wall pattern in metaxylem vessels is specified by transcriptional control of actin polymerization. A newly isolated allele of KNOTTED-LIKE HOMEOBOX TRANSCRIPTION FACTOR 7 (KNAT7) was associated with the formation of banded cell walls in metaxylem vessels. Loss of KNAT7 caused misexpression of FORMIN HOMOLOGY DOMAIN CONTAINING PROTEIN11 (FH11) in the metaxylem, which in turn caused rearrangements of ROP GTPases and microtubules in banded patterns. FH11 function required its plasma membrane anchoring and actin polymerization activity. These results suggest that excessive actin polymerization at the plasma membrane abolishes the pitted cell wall formation and promotes banded cell wall formation in metaxylem vessels. This study unveils the importance of proper control of actin polymerization for cell wall pattern determination.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Zhong, R. Q., Cui, D. T. & Ye, Z. H. Secondary cell wall biosynthesis. New Phytologist221, 1703–1723 (2019). - PubMed
-
- Oda, Y. & Fukuda, H. Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol.15, 38–44 (2012). - PubMed
-
- Watanabe, Y. et al. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science350, 198–203 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

