PBRM1 directs PBAF to pericentromeres and protects centromere integrity
- PMID: 40011561
- PMCID: PMC11865495
- DOI: 10.1038/s41467-025-57277-9
PBRM1 directs PBAF to pericentromeres and protects centromere integrity
Abstract
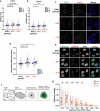
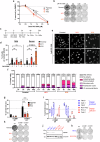
The specialised structure of the centromere is critical for effective chromosome segregation, but its repetitive nature makes it vulnerable to rearrangements. Centromere fragility can drive tumorigenesis, but protective mechanisms preventing fragility are still not fully understood. The PBAF chromatin remodelling complex is frequently misregulated in cancer, but its role in cancer is incompletely characterized. Here, we identify PBAF as a protector of centromere and pericentromere structure with profound consequences for genome stability. A conserved feature of isogenic cell lines lacking PBRM1, a subunit of PBAF, is compromised centromere and pericentromere integrity. PBAF is present at these regions, and binding patterns of PBAF and H3K9 methylation change when PBRM1 is absent. PBRM1 loss creates a dependence on the spindle assembly checkpoint, which represents a therapeutic vulnerability. Importantly, we find that even in the absence of any perturbations, PBRM1 loss leads to centromere fragility, thus identifying a key player in centromere protection.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

