Local structural flexibility drives oligomorphism in computationally designed protein assemblies
- PMID: 40011747
- PMCID: PMC12170342
- DOI: 10.1038/s41594-025-01490-z
Local structural flexibility drives oligomorphism in computationally designed protein assemblies
Abstract
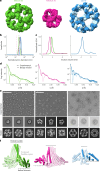
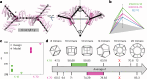
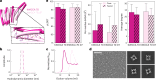
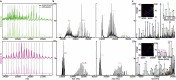
Many naturally occurring protein assemblies have dynamic structures that allow them to perform specialized functions. Although computational methods for designing novel self-assembling proteins have advanced substantially over the past decade, they primarily focus on designing static structures. Here we characterize three distinct computationally designed protein assemblies that exhibit unanticipated structural diversity arising from flexibility in their subunits. Cryo-EM single-particle reconstructions and native mass spectrometry reveal two distinct architectures for two assemblies, while six cryo-EM reconstructions for the third likely represent a subset of its solution-phase structures. Structural modeling and molecular dynamics simulations indicate that constrained flexibility within the subunits of each assembly promotes a defined range of architectures rather than nonspecific aggregation. Redesigning the flexible region in one building block rescues the intended monomorphic assembly. These findings highlight structural flexibility as a powerful design principle, enabling exploration of new structural and functional spaces in protein assembly design.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors have patent filings to disclose: J.Y.(J.)W., A.K., D.B. and N.P.K. are coinventors on patent applications (International patent application PCT/US2023/065397 (2023) and US provisional patent application 63/328,394 (2022)) related to this work. The other authors declare no competing interests.
Figures








Update of
-
Local structural flexibility drives oligomorphism in computationally designed protein assemblies.bioRxiv [Preprint]. 2023 Oct 19:2023.10.18.562842. doi: 10.1101/2023.10.18.562842. bioRxiv. 2023. Update in: Nat Struct Mol Biol. 2025 Jun;32(6):1050-1060. doi: 10.1038/s41594-025-01490-z. PMID: 37905007 Free PMC article. Updated. Preprint.
References
-
- Howorka, S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr. Opin. Biotechnol.22, 485–491 (2011). - PubMed
-
- Lee, E. J., Lee, N. K. & Kim, I.-S. Bioengineered protein-based nanocage for drug delivery. Adv. Drug Deliv. Rev.106, 157–171 (2016). - PubMed
-
- Basu Mallik, B., Stanislaw, J., Alawathurage, T. M. & Khmelinskaia, A. De novo design of polyhedral protein assemblies: before and after the AI revolution. Chem. Bio. Chem.24, e202300117 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

