Hsa_circ_0125356 promotes gemcitabine resistance by modulating WNT canonical and non-canonical pathways via miR-582-5p/FGF9 axis in non-small cell lung cancer
- PMID: 40011917
- PMCID: PMC11866803
- DOI: 10.1186/s12943-025-02259-0
Hsa_circ_0125356 promotes gemcitabine resistance by modulating WNT canonical and non-canonical pathways via miR-582-5p/FGF9 axis in non-small cell lung cancer
Abstract
Background: Non-small cell lung cancer (NSCLC) is the leading cause of cancer morbidity and mortality worldwide. The prognosis of patients has been significantly improved by chemotherapy, but acquired drug resistance remains a major obstacle to NSCLC treatment. Circular RNAs (circRNAs), which act as miRNA or protein sponges, are critically associated with the development and chemotherapy resistance of NSCLC.
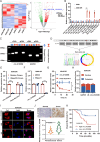
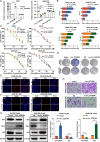
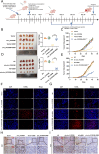
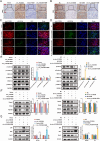
Methods: CircRNA sequencing was performed to analyze the differential expression of circRNAs between A549 and A549-GR cells. Chromogenic in situ hybridization (CISH) and immunohistochemistry (IHC) technologies were used to detect the expression of hsa_circ_0125356, miR-582-5p,and FGF9 in NSCLC tissues and para-carcinoma tissues. Fluorescence in situ hybridization (FISH), dual-luciferase reporter assays and RNA immunoprecipitation (RIP) were conducted to evaluate the expression and regulation of hsa_circ_0125356, miR-582-5p, and FGF9. Furthermore, the regulation of hsa_circ_0125356/miR-582-5p/FGF9 on gemcitabine sensitivity was confirmed by TUNEL, Transwell, EdU, CCK8 and immunohistochemistry.
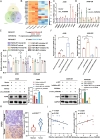
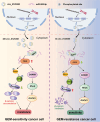
Results: We identified a novel hsa_circ_0125356 as a therapeutic target against gemcitabine resistance. Hsa_circ_0125356 was significantly elevated in clinical samples of patients with NSCLC. Moreover, hsa_circ_0125356 overexpression promoted gemcitabine resistance to NSCLC by upregulating FGF9 via sponging miR-582-5p in vivo and in vitro. Notably, WNT canonical (ERK/GSK3β/β-catenin) and non-canonical (Daam1/RhoA/ROCK2) signaling pathways were activated due to hsa_circ_0125356 acting as an endogenous miR-582-5p sponge to regulate the expression of FGF9, and thereby enhancing gemcitabine resistance via promoting DNA damage repair and inhibition of apoptosis. The results were further confirmed by two small molecule antagonists, WAY 316606 and XAV-939,which could inhibit the activation of WNT signaling pathway induced by hsa_circ_0125356.
Conclusion: We first demonstrated that hsa_circ_0125356 was significantly upregulated and served as a biomarker for gemcitabine resistance in NSCLC by sponging miR-582-5p/FGF9 axis to regulate the WNT canonical and non-canonical signaling pathways, which provided a new direction for identification of therapeutic targets for the treatment of gemcitabine resistance of NSCLC.
Keywords: CircRNAs; FGF9; Gemcitabine resistance; MiR-582-5p; NSCLC; WNT canonical and non-canonical signaling pathways.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: All authors agreed to the publication of the manuscript. Competing interests: The authors declare no competing interests.
Figures




References
-
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53. - PubMed
-
- Del VV, Denti MA. microRNA and Lung Cancer. Adv Exp Med Biol. 2015;889:153–77. - PubMed
-
- Seidl C, Panzitt K, Bertsch A, Brcic L, Schein S, Mack M, Leithner K, Prinz F, Olschewski H, Kornmueller K, Hrzenjak A. MicroRNA-182-5p regulates hedgehog signaling pathway and chemosensitivity of cisplatin-resistant lung adenocarcinoma cells via targeting GLI2. Cancer Lett. 2020;469:266–76. - PubMed
-
- Zhong L, Sun S, Shi J, Cao F, Han X, Chen Z. MicroRNA-125a-5p plays a role as a tumor suppressor in lung carcinoma cells by directly targeting STAT3. Tumour Biol. 2017;39:1393392085. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

