IFNλ1 is a STING-dependent mediator of DNA damage and induces immune activation in lung cancer
- PMID: 40012911
- PMCID: PMC11862833
- DOI: 10.3389/fimmu.2024.1525083
IFNλ1 is a STING-dependent mediator of DNA damage and induces immune activation in lung cancer
Abstract
Introduction: The importance of the cGAS-STING pathway and type I interferon (IFN) in anti-tumor immunity has been widely studied. However, there is limited knowledge about the role of type III IFNs in cancer settings. Type III IFNs, comprising IFNλ1-4, are opposite to type I IFN only expressed by a few cell types, including epithelial cells, and the receptor subunit IFNLR1, is equally only expressed on limited types of cells.
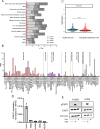
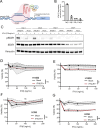
Methods: Gene and protein expression of the cGAS-STING signaling pathway was characterized in a series of non-small cell lung cancer (NSCLC) cell lines. Herring-testis DNA stimulation and chemotherapy drugs (doxorubicin and cisplatin) were used to activate the cGAS-STING pathway, and the level of activation was determined by measuring changes in the transcriptomic profile as well as type I and III IFNs by ELISA. Re-expression of IFNLR1 on cancer cell lines was achieved using CRISPR activation (CRISPRa) followed by evaluating chemotherapy-induced apoptosis using flow cytometry assays.
Results: STING was not broadly expressed across the NSCLC cell lines. Those cancer cell lines expressing all relevant factors supporting the cGAS-STING pathway secreted IFNλ following STING activation whereas only few of them expressed IFNβ. Treatment with chemotherapy drugs likewise preferentially induced IFNλ, which was abrogated in CRISPR-Cas9 STING knock-out cells. Expression of IFNLR1 was found downregulated in the cancer cell lines compared to the benign epithelial cell line Nuli-1. Rescuing IFNLR1 expression by CRISPRa in multiple cancer cell lines sensitization them to IFNλ-stimulation and resulted in significant reduction in cell viability.
Conclusion: Downregulation of IFNLR1 can be an immune evasion mechanism developed by cancer cells to avoid responding to endogenous type III IFNs. Thus, rescuing IFNLR1 expression in NSCLC in conjunction to chemotherapy may potentially be harnessed to elevate the anti-tumoral responses.
Keywords: CRISPR/Cas9; NSCLC; STING; cancer immunology; interferon lambda.
Copyright © 2025 Godsk, Jensen, Larsen, Ahrenfeldt, Gammelgaard and Jakobsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Kumar A, Taghi Khani A, Duault C, Aramburo S, Sanchez Ortiz A, Lee SJ, et al. . Intrinsic suppression of type I interferon production underlies the therapeutic efficacy of IL-15-producing natural killer cells in B-cell acute lymphoblastic leukemia. J Immunother Cancer. (2023) 11(5). doi: 10.1136/jitc-2022-006649 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

