A Lgr5-independent developmental lineage is involved in mouse intestinal regeneration
- PMID: 40013494
- PMCID: PMC12045596
- DOI: 10.1242/dev.204654
A Lgr5-independent developmental lineage is involved in mouse intestinal regeneration
Abstract
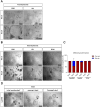
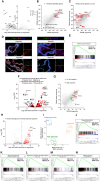
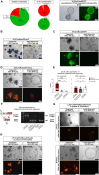
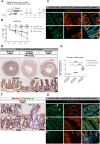
Collagenase and dispase treatment of intestinal tissue from adult mice generates cells growing in matrigel as stably replatable cystic spheroids, in addition to differentiated organoids. Contrary to classical EDTA-derived organoids, these spheroids display poor intestinal differentiation and grow independently of Rspondin, noggin and EGF. Their transcriptome strikingly resembles that of fetal intestinal spheroids, with downregulation of crypt base columnar cell (CBC) markers (Lgr5, Ascl2, Smoc2 and Olfm4). In addition, they display upregulation of inflammatory and mesenchymal genetic programs, together with robust expression of YAP target genes. Lineage tracing, cell-sorting and single cell RNA sequencing experiments demonstrate that adult spheroid-generating cells belong to a hitherto undescribed developmental lineage, independent of Lgr5-positive CBCs, and are involved in regeneration of the epithelium following CBC ablation.
Keywords: Intestinal development; Organoids; Regeneration; Stem cells.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

