Design and synthesis of novel HDAC6 inhibitor dimer as HDAC6 degrader for cancer treatment by palladium catalysed dimerisation
- PMID: 40013582
- PMCID: PMC11869342
- DOI: 10.1080/14756366.2025.2468355
Design and synthesis of novel HDAC6 inhibitor dimer as HDAC6 degrader for cancer treatment by palladium catalysed dimerisation
Abstract
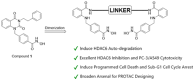
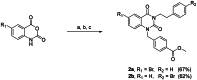
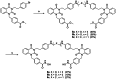
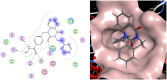
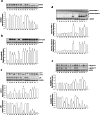
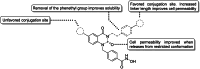
The enigmatic histone deacetylase 6 (HDAC6) is one of a kind among its family. Recent reports revealed that HDAC6 CD1 exhibits E3 ligase activity. Inspired by these researches, we attempted to develop drugs targeting HDAC6 via novel mechanism. Herein, we report a palladium catalysed transformation and purification method for hydroxamic acid dimers, and series of HDAC6 inhibitor-based dimer showing outstanding biological activities and capability of inducing auto-degradation. Our proof-of-concept was highlighted with 2-amino benzamide-based HDAC6 inhibitor dimers that exhibit great HDAC6 inhibition activity (3.9-15.4 nM), good HDAC1/6 selectivity (95-577), and excellent cytotoxicity against human hormone-resistant prostate cancer (HRPC) PC-3 and non-small cell lung cancer (NSCLC) A549 cell lines (5.9-11.3 and 6.6-17.9 μM, respectively) while simultaneously inducing HDAC6 degradation. These dimers not only induce apoptosis and autophagy but also interfere with kinetochore attachment by the detection of BUBR1 phosphorylation at S670.
Keywords: HDAC6; anticancer; coupling; palladium; protein degrader.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures






References
-
- Gregoretti IV, Lee YM, Goodson HV.. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. - PubMed
-
- Pulya S, Amin SA, Adhikari N, Biswas S, Jha T, Ghosh B.. Hdac6 as privileged target in drug discovery: a perspective. Pharmacol Res. 2021;163:105274. - PubMed
-
- Kalin JH, Bergman JA.. Development and therapeutic implications of selective histone deacetylase 6 inhibitors. J Med Chem. 2013;56(16):6297–6313. - PubMed
-
- Liu Y, Li L, Min J.. Hdac6 finally crystal clear. Nat Chem Biol. 2016;12(10):885–885. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
