Rho-dependent termination and RNase E-mediated cleavage: dual pathways for RNA 3' end processing in polycistronic mRNA
- PMID: 40013805
- PMCID: PMC11925234
- DOI: 10.1128/jb.00437-24
Rho-dependent termination and RNase E-mediated cleavage: dual pathways for RNA 3' end processing in polycistronic mRNA
Abstract
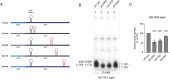
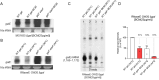
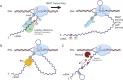
"Pre-full-length" transcripts are produced at the end of the polycistronic galactose (gal) operon, 5' galE-galT-galK-galM 3', via Rho-dependent transcription termination (RDT) and -independent transcription termination. The 3' end of the full-length galETKM mRNA is acquired by exonucleolytic processing of the 3'-OH ends of the pre-full-length transcripts. However, the gal operon produces an mRNA termed galE whose 3' end forms approximately 120 nucleotides downstream of the galE stop codon, within the subsequent gene, galT, thereby establishing polarity in gene expression. In this study, we investigated the molecular processes that generate the 3' end of galE mRNA. We discovered that the 3' ends of pre-galE mRNA are produced in the middle of galT as a result of the combination of two separate molecular processes-one previously reported as RDT and the other as unreported RNase E-mediated transcript cleavage. The 3' ends of pre-galE mRNA undergo exonucleolytic processing to the 3' end of galE mRNA observed in vivo. A hairpin structure containing an 8 bp stem and a 4-nucleotide loop, located 5-10 nucleotides upstream of the 3' ends of galE mRNA, blocks exoribonuclease digestion and renders transcript stability. These findings demonstrate that RNase E-contrary to its general role in mRNA degradation-produces RNA 3' ends that regulate polarity in gene expression.IMPORTANCEThis study reports the findings of two molecular mechanisms that generate the 3' ends of pre-galE mRNA in the gal operon, viz., Rho-dependent transcription termination and RNase E-mediated cleavage. These 3' ends are subsequently processed to produce stable galE mRNA with a hairpin structure that prevents exoribonuclease degradation. This mechanism establishes gene expression polarity by generating the 3' end of galE mRNA within galT in contrast to the usual mRNA degradation role of RNase E. The study reveals a unique role of RNase E in mRNA processing and stability.
Keywords: RNase E-mediated cleavage; Rho-dependent transcription termination; exoribonuclease digestion; mRNA 3′ end; polarity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

