Flavonoid-converting capabilities of Clostridium butyricum
- PMID: 40014075
- PMCID: PMC11868195
- DOI: 10.1007/s00253-025-13434-0
Flavonoid-converting capabilities of Clostridium butyricum
Abstract
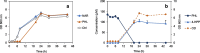
Clostridium butyricum inhabits various anoxic environments, including soil and the human gut. Here, this common bacterium comes into contact with abundant plant-derived flavonoids. Metabolization of these bioactive polyphenols has been studied in recent years, particularly focusing on gut bacteria due to the proposed health-promoting properties of these dietary constituents. Based on an initial report in 1997 on eriodictyol degradation (Miyake et al. 1997, J Agric Food Chem, 45:3738-3742), the present study systematically investigated C. butyricum for its ability to convert a set of structurally diverse flavonoids. Incubation experiments revealed that C. butyricum deglycosylated flavonoid O-glucosides but only when glucose was absent. Moreover, aglycone members of flavone, flavanone, dihydrochalcone, and flavanonol subclasses were degraded. The C-ring cleavage of the flavanones, naringenin and eriodictyol, was stereospecific and finally resulted in formation of the corresponding hydroxyphenylpropionic acids. Stereospecific C-ring cleavage of the flavanonol taxifolin led to taxifolin dihydrochalcone. C. butyricum did neither cleave flavonols and isoflavones nor catalyze de-rhamnosylation, demethylation, or dehydroxylation of flavonoids. Genes encoding potential flavonoid-metabolizing enzymes were detected in the C. butyricum genome. Overall, these findings indicate that C. butyricum utilizes flavonoids as alternative substrates and, as observed for the dihydrochalcone phloretin, can eliminate growth-inhibiting flavonoids through degradation. KEY POINTS: • Clostridium butyricum deglycosylated flavonoid O-glucosides. • Clostridium butyricum converted members of several flavonoid subclasses. • Potential flavonoid-metabolizing enzymes are encoded in the C. butyricum genome.
Keywords: Clostridium butyricum; Flavonoid; Flavonoid glycoside; Gut bacterium; Polyphenol; Soil bacterium.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: This article does not contain any studies involving human participants or animals performed by the author. Conflict of interest: The author declares no competing interests.
Figures

References
-
- Barreca D, Bellocco E, Lagana G, Ginestra G, Bisignano C (2014) Biochemical and antimicrobial activity of phloretin and its glycosylated derivatives present in apple and kumquat. Food Chem 160:292–297. 10.1016/j.foodchem.2014.03.118 - PubMed
-
- Braune A, Blaut M (2011) Deglycosylation of puerarin and other aromatic C-glucosides by a newly isolated human intestinal bacterium. Environ Microbiol 13:482–494. 10.1111/j.1462-2920.2010.02352.x - PubMed
-
- Braune A, Blaut M (2018) Catenibacillus scindens gen. nov., sp. nov., a C-deglycosylating human intestinal representative of the Lachnospiraceae. Int J Syst Evol Microbiol 68:3356–3361. 10.1099/ijsem.0.003001 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

