Decoding functional hematopoietic progenitor cells in the adult human lung
- PMID: 40014797
- PMCID: PMC7617544
- DOI: 10.1182/blood.2024027884
Decoding functional hematopoietic progenitor cells in the adult human lung
Abstract
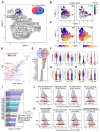
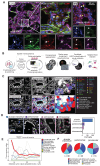
Although the bone marrow is the main site of blood cell production in adults, rare pools of hematopoietic stem and progenitor cells have been found in extramedullary organs. In mice, we have previously shown that the lung contains hematopoietic progenitor cells and is a site of platelet production. Here, in the adult human lung, we show that functional hematopoietic precursors reside in the extravascular spaces with a frequency similar to the bone marrow and are capable of proliferation and engraftment in mice. The gene signature of pulmonary and medullary CD34+ hematopoietic progenitors indicates greater baseline activation of immune-, megakaryocyte/platelet-, and erythroid-related pathways in lung progenitors. Spatial transcriptomics mapped blood progenitors in the lung to an alveolar interstitium niche with only a few cells identified in an intravascular location. In human blood samples collected for stem cell transplantation, CD34+ cells with a lung signature enriched the mobilized pool of hematopoietic stem cells. These results identify the lung as a pool for uniquely programmed blood stem and progenitor cells with the potential to support hematopoiesis in humans.
© 2025 American Society of Hematology. Published by Elsevier Inc. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Conflict of interest statement
Figures



Update of
-
Decoding functional hematopoietic progenitor cells in the adult human lung.Res Sq [Preprint]. 2024 Feb 16:rs.3.rs-3576483. doi: 10.21203/rs.3.rs-3576483/v2. Res Sq. 2024. Update in: Blood. 2025 May 1;145(18):1975-1986. doi: 10.1182/blood.2024027884. PMID: 38077002 Free PMC article. Updated. Preprint.
Comment in
-
Breathing life into the hematopoietic potential of the lung.Blood. 2025 May 1;145(18):1963-1964. doi: 10.1182/blood.2025028548. Blood. 2025. PMID: 40310663 No abstract available.
References
-
- Abkowitz JL, Catlin SN, McCallie MT, et al. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood. 2002;100:2665–2667. - PubMed

