Probing the Dynamics of Yersinia Adhesin A (YadA) in Outer Membranes Hints at Requirements for β-Barrel Membrane Insertion
- PMID: 40014811
- PMCID: PMC11912334
- DOI: 10.1021/jacs.4c17726
Probing the Dynamics of Yersinia Adhesin A (YadA) in Outer Membranes Hints at Requirements for β-Barrel Membrane Insertion
Abstract
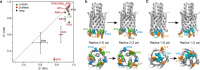
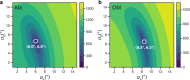
The vast majority of cells are protected and functionalized by a dense surface layer of glycans, proteoglycans, and glycolipids. This surface represents an underexplored space in structural biology that is exceedingly challenging to recreate in vitro. Here, we investigate β-barrel protein dynamics within an asymmetric outer membrane environment, with the trimeric autotransporter Yersinia adhesin A (YadA) as an example. Magic-angle spinning NMR relaxation data and a model-free approach reveal increased mobility in the second half of strand β2 after the conserved G72, which is responsible for membrane insertion and autotransport, and in the subsequent loop toward β3. In contrast, the protomer-protomer interaction sites (β1i-β4i-1) are rigid. Intriguingly, the mobility in the β-strand section following G72 is substantially elevated in the outer membrane and less so in the detergent environment of microcrystals. A possible source is revealed by molecular dynamics simulations that show the formation of a salt bridge involving E79 and R76 in competition with a dynamic interplay of calcium binding by E79 and the phosphate groups of the lipids. An estimation of overall barrel motion in the outer membrane and detergent-containing crystals yields values of around 41 ns for both. The global motion of YadA in the outer membrane has a stronger rotational component orthogonal to the symmetry axis of the trimeric porin than in the detergent-containing crystal. In summary, our investigation shows that the mobility in the second half of β2 and the loop to β3 required for membrane insertion and autotransport is maintained in the final folded form of YadA.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

