p85β acts as a transcription cofactor and cooperates with BCLAF1 in the nucleus
- PMID: 40016211
- PMCID: PMC11868507
- DOI: 10.1038/s41467-025-56532-3
p85β acts as a transcription cofactor and cooperates with BCLAF1 in the nucleus
Abstract
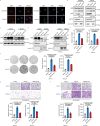
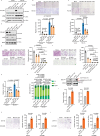
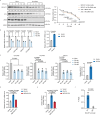
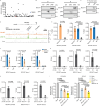
p85β is a regulatory subunit of the phosphoinositide 3-kinase (PI3K). Emerging evidence suggests that p85β goes beyond its role in the PI3K and is functional in the nucleus. In this study, we discover that nuclear p85β is enriched at gene loci and regulates gene transcription and that this regulatory role contributes to the oncogenic potential of nuclear p85β. A multi-omics approach reveals the physical interaction and functional cooperativity between nuclear p85β and a transcription factor BCLAF1. We observe genome-wide co-occupancy of p85β and BCLAF1 at gene targets associated with transcriptional responses. Intriguingly, the targetome includes BCLAF1 of which transcription is activated by p85β and BCLAF1, indicating a positive autoregulation. While BCLAF1 recruits p85β to BCLAF1 loci, p85β facilitates the assembly of BCLAF1, the scaffold protein TRIM28 and the zinc finger transcription factor ZNF263, which together act in concert to activate BCLAF1 transcription. Collectively, this study provides functional evidence and mechanistic basis to support a role of nuclear p85β in modulating gene transcription.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

