Circulating miR-126-3p is a mechanistic biomarker for knee osteoarthritis
- PMID: 40016267
- PMCID: PMC11868599
- DOI: 10.1038/s41467-025-57308-5
Circulating miR-126-3p is a mechanistic biomarker for knee osteoarthritis
Abstract
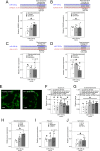
Osteoarthritis is a major contributor to pain and disability worldwide, yet there are currently no validated soluble biomarkers or disease-modifying treatments. Given that microRNAs are promising mechanistic biomarkers that can be therapeutically targeted, in this study, we aimed to identify and prioritize reproducible circulating microRNAs associated with radiographic knee osteoarthritis. Across four independent cohorts, we find circulating miR-126-3p is elevated in knee osteoarthritis versus controls. Across six primary human knee osteoarthritis tissues, miR-126-3p is highest in subchondral bone, fat pad and synovium, and lowest in cartilage. Following both intravenous and intra-articular miR-126-3p mimic treatment in a surgical mouse model of knee osteoarthritis, we show reduced disease severity in males. In human knee osteoarthritis biospecimens, miR-126-3p mimic treatment reduces genes and markers associated with angiogenesis, as well as genes linked to osteogenesis, adipogenesis, and synovitis-processes secondary to angiogenesis. Our findings indicate that miR-126-3p is elevated in knee osteoarthritis and mitigates disease severity, supporting its potential as a biomarker and therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Buckwalter, J. A. & Mankin, H. J. Articular cartilage: degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr. Course Lect.47, 487–504 (1998). - PubMed
-
- Rocha, F. A. C. & Ali, S. A. Soluble biomarkers in osteoarthritis in 2022: year in review. Osteoarthr. Cartil.31, 167–176 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

