International Precision Child Health Partnership (IPCHiP): an initiative to accelerate discovery and improve outcomes in rare pediatric disease
- PMID: 40016282
- PMCID: PMC11868529
- DOI: 10.1038/s41525-025-00474-8
International Precision Child Health Partnership (IPCHiP): an initiative to accelerate discovery and improve outcomes in rare pediatric disease
Abstract
Advances in genomic technologies have revolutionized the diagnosis of rare genetic diseases, leading to the emergence of precision therapies. However, there remains significant effort ahead to ensure the promise of precision medicine translates to improved outcomes. Here, we discuss the challenges in advancing precision child health and highlight how international collaborations such as the International Precision Child Health Partnership, which embed research into clinical care, can maximize benefits for children globally.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.B.H. has received research funding from UCB Australia, Praxis Precision Medicines and RogCon Biosciences, Inc., has served on an advisory board for UCB Australia, and is a member of the Scientific and Medical Board for SCN2A Asia-Pacific. N.C.A. is on the Boards of Directors of Novartis, Charles River Laboratories and Maze Therapeutics, and the Scientific Advisory Board of Dyne Therapeutics. A.H.B. has received consulting fees from Astellas Gene Therapies, GLG Inc., Guidepoint Global, and F. Hoffman-La Roche, is on the Scientific Advisory Board of Kate Therapeutics, and holds equity in Kate Therapeutics and Kinea Bio. J.H.C. has acted as an investigator for studies with GW Pharma/Jazz Pharmaceuticals, Zogenix/UCB Pharma, Vitaflo, Stoke Therapeutics, and Ultragenyx. She has been a speaker and on advisory boards for Jazz Pharmaceuticals, UCB, Biocodex, and Nutricia; all remuneration has been paid to her department. J.C. is a member of the Drug Monitoring and Safety Board for Anavex Life Sciences Corp, the Endpoint Adjudication Committee for Taysha Gene Therapies, the Scientific and Medical Advisory Committees of the Childhood Dementia Initiative and the Rett Syndrome Association of Australia, and is the director of the Mito Foundation of Australia. S.W.S. served on the scientific advisory committee of Population Bio, Deep Genomics and intellectual property from his research held at the Hospital for Sick Children and licensed to Athena Diagnostics and Population Bio. S.W.S., R.D.C., and C.R.M. are Editors or Editorial Board Members of npj Genomic Medicine; they were not part of the peer review or decision-making processes for this manuscript. S.M.W., A.McT., A.M.D., G.C., A.P., I.E.S., V.C., L.D.S., S.E.M.S., M.W., A.D., N.S., P.S., L.S.C., and K.N.N. report no competing interests.
Figures
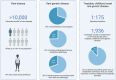




References
-
- Richter, T. et al. Rare disease terminology and definitions-a systematic global review: report of the ISPOR rare disease special interest group. Value Health18, 906–914 (2015). - PubMed
-
- Ferreira, C. R. The burden of rare diseases. Am. J. Med. Genet. A179, 885–892 (2019). - PubMed
-
- Dodge, J. A. et al. The importance of rare diseases: from the gene to society. Arch. Dis. Child.96, 791–792 (2011). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

