Nasal anti-CD3 monoclonal antibody ameliorates traumatic brain injury, enhances microglial phagocytosis and reduces neuroinflammation via IL-10-dependent Treg-microglia crosstalk
- PMID: 40016353
- PMCID: PMC11893472
- DOI: 10.1038/s41593-025-01877-7
Nasal anti-CD3 monoclonal antibody ameliorates traumatic brain injury, enhances microglial phagocytosis and reduces neuroinflammation via IL-10-dependent Treg-microglia crosstalk
Abstract
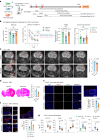
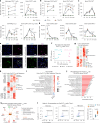
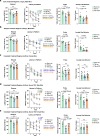
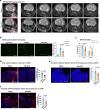
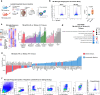
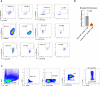
Neuroinflammation plays a crucial role in traumatic brain injury (TBI), contributing to both damage and recovery, yet no effective therapy exists to mitigate central nervous system (CNS) injury and promote recovery after TBI. In the present study, we found that nasal administration of an anti-CD3 monoclonal antibody ameliorated CNS damage and behavioral deficits in a mouse model of contusional TBI. Nasal anti-CD3 induced a population of interleukin (IL)-10-producing regulatory T cells (Treg cells) that migrated to the brain and closely contacted microglia. Treg cells directly reduced chronic microglia inflammation and regulated their phagocytic function in an IL-10-dependent manner. Blocking the IL-10 receptor globally or specifically on microglia in vivo abrogated the beneficial effects of nasal anti-CD3. However, the adoptive transfer of IL-10-producing Treg cells to TBI-injured mice restored these beneficial effects by enhancing microglial phagocytic capacity and reducing microglia-induced neuroinflammation. These findings suggest that nasal anti-CD3 represents a promising new therapeutic approach for treating TBI and potentially other forms of acute brain injury.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.L.W. is on the scientific advisory board of Tiziana Life Sciences.
Figures











References
-
- Faul, M. & Coronado, V. Epidemiology of traumatic brain injury. Handb. Clin. Neurol.127, 3–13 (2015). - PubMed
-
- Centers for Disease Control and Prevention. Surveillance Report of Traumatic Brain Injury-related Emergency Department Visits, Hospitalizations, and Deaths—United States, 2014 (CDC, 2019).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

