Epidemiological Dynamics in Populations Structured by Neighbourhoods and Households
- PMID: 40016448
- PMCID: PMC11868190
- DOI: 10.1007/s11538-025-01426-0
Epidemiological Dynamics in Populations Structured by Neighbourhoods and Households
Abstract
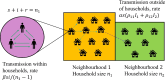
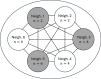
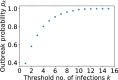
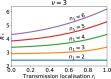
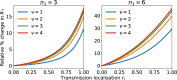
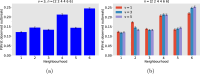
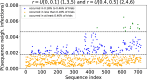
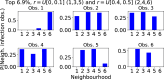
Epidemiological dynamics are affected by the spatial and demographic structure of the host population. Households and neighbourhoods are known to be important groupings but little is known about the epidemiological interplay between them. In order to explore the implications for infectious disease epidemiology of households with similar demographic structures clustered in space we develop a multi-scale epidemic model consisting of neighbourhoods of households. In our analysis we focus on key parameters which control household size, the importance of transmission within households relative to outside of them, and the degree to which the non-household transmission is localised within neighbourhoods. We construct the household reproduction number over all neighbourhoods and derive the analytic probability of an outbreak occurring from a single infected individual in a specific neighbourhood. We find that reduced localisation of transmission within neighbourhoods reduces when household size differs between neighbourhoods. This effect is amplified by larger differences between household sizes and larger divergence between transmission rates within households and outside of them. However, the impact of neighbourhoods with larger household sizes on an individual's risk of infection is mainly limited to the individuals that reside in those neighbourhoods. We consider various surveillance scenarios and show that household size information from the initial infectious cases is often more important than neighbourhood information while household size and neighbourhood localisation influences the sequence of neighbourhoods in which an outbreak is observed.
Keywords: Epidemiology; Household; Mathematical model; Metapopulation; Neighbourhood; Outbreak probability; Reproduction number; Surveillance.
© 2025. The Author(s).
Figures



Similar articles
-
Effects of pathogen dependency in a multi-pathogen infectious disease system including population level heterogeneity - a simulation study.Theor Biol Med Model. 2017 Dec 13;14(1):26. doi: 10.1186/s12976-017-0072-7. Theor Biol Med Model. 2017. PMID: 29237462 Free PMC article.
-
Estimating the within-household infection rate in emerging SIR epidemics among a community of households.J Math Biol. 2015 Dec;71(6-7):1705-35. doi: 10.1007/s00285-015-0872-5. Epub 2015 Mar 28. J Math Biol. 2015. PMID: 25820343
-
Stochastic SIR epidemics in a population with households and schools.J Math Biol. 2016 Apr;72(5):1177-93. doi: 10.1007/s00285-015-0901-4. Epub 2015 Jun 13. J Math Biol. 2016. PMID: 26070348 Free PMC article.
-
Traveling waves for an epidemic patchy model with bilinear incidence.J Math Biol. 2025 May 21;90(6):63. doi: 10.1007/s00285-025-02228-7. J Math Biol. 2025. PMID: 40397138
-
Evaluation of vaccination strategies for SIR epidemics on random networks incorporating household structure.J Math Biol. 2018 Jan;76(1-2):483-530. doi: 10.1007/s00285-017-1139-0. Epub 2017 Jun 20. J Math Biol. 2018. PMID: 28634747 Free PMC article.
References
-
- Adams B (2016) Household demographic determinants of Ebola epidemic risk. J Theor Biol 392:99–106 - PubMed
-
- Athreya K, Ney P (1972) Branching processes. New York
-
- Ball F, Lyne OD (2001) Stochastic multi-type SIR epidemics among a population partitioned into households. Adv Appl Probab 33(1):99–123
-
- Ball F, Neal P (2002) A general model for stochastic SIR epidemics with two levels of mixing. Math Biosci 180(1–2):73–102 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

