Autophagy repression by antigen and cytokines shapes mitochondrial, migration and effector machinery in CD8 T cells
- PMID: 40016525
- PMCID: PMC11876071
- DOI: 10.1038/s41590-025-02090-1
Autophagy repression by antigen and cytokines shapes mitochondrial, migration and effector machinery in CD8 T cells
Abstract
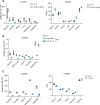
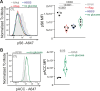
Autophagy shapes CD8 T cell fate; yet the timing, triggers and targets of this process are poorly defined. Herein, we show that naive CD8 T cells have high autophagic flux, and we identify an autophagy checkpoint whereby antigen receptor engagement and inflammatory cytokines acutely repress autophagy by regulating amino acid transporter expression and intracellular amino acid delivery. Activated T cells with high levels of amino acid transporters have low autophagic flux in amino-acid-replete conditions but rapidly reinduce autophagy when amino acids are restricted. A census of proteins degraded and fueled by autophagy shows how autophagy shapes CD8 T cell proteomes. In cytotoxic T cells, dominant autophagy substrates include cytolytic effector molecules, and amino acid and glucose transporters. In naive T cells, mitophagy dominates and selective mitochondrial pruning supports the expression of molecules that coordinate T cell migration and survival. Autophagy thus differentially prunes naive and effector T cell proteomes and is dynamically repressed by antigen receptors and inflammatory cytokines to shape T cell differentiation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Jia, W. & He, Y.-W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J. Immunol.186, 5313–5322 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

