DOG1 controls dormancy independently of ABA core signaling kinases regulation by preventing AFP dephosphorylation through AHG1
- PMID: 40020062
- PMCID: PMC11870083
- DOI: 10.1126/sciadv.adr8502
DOG1 controls dormancy independently of ABA core signaling kinases regulation by preventing AFP dephosphorylation through AHG1
Abstract
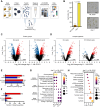
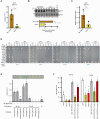
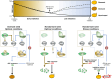
Seed dormancy determines germination timing, influencing seed plant adaptation and overall fitness. DELAY OF GERMINATION 1 (DOG1) is a conserved central regulator of dormancy cooperating with the phytohormone abscisic acid (ABA) through negative regulation of ABA HYPERSENSITIVE GERMINATION (AHG) 1 and AHG3 phosphatases. The current molecular mechanism of DOG1 signaling proposes it regulates the activation of central ABA-related SnRK2 kinases. Here, we unveil DOG1's functional autonomy from the regulation of ABA core signaling components and unravel its pivotal control over the activation of ABSCISIC ACID INSENSITIVE FIVE BINDING PROTEINs (AFPs). Our data revealed a molecular relay in which AFPs' genuine activation by AHG1 is contained by DOG1 to prevent the breakdown of maturation-imposed ABA responses independently of ABA-related kinase activation status. This work offers a molecular understanding of how plants fine-tune germination timing, while preserving seed responsiveness to adverse environmental cues, and thus represents a milestone in the realm of conservation and breeding programs.
Figures


References
-
- C. Baroux, U. Grossniklaus, “Chapter twenty-one - Seeds—An evolutionary innovation underlying reproductive success in flowering plants” in Current Topics in Developmental Biology (Academic Press, 2019), vol. 131, pp. 605–642. - PubMed
-
- de Vries J., Archibald J. M., Plant evolution: Landmarks on the path to terrestrial life. New Phytol. 217, 1428–1434 (2018). - PubMed
-
- J. D. Bewley, M. Black, Physiology and Biochemistry of Seeds in Relation to Germination: Volume 2: Viability, Dormancy, and Environmental Control (Springer, 2012).
-
- Finch-Savage W. E., Leubner-Metzger G., Seed dormancy and the control of germination. New Phytol. 171, 501–523 (2006). - PubMed
-
- Née G., Xiang Y., Soppe W. J. J., The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

