A long-lived pool of PINK1 imparts a molecular memory of depolarization-induced activity
- PMID: 40020067
- PMCID: PMC11870087
- DOI: 10.1126/sciadv.adr1938
A long-lived pool of PINK1 imparts a molecular memory of depolarization-induced activity
Abstract
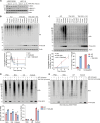
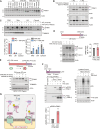
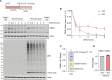
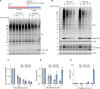
The Parkinson's disease-linked kinase, PINK1, is a short-lived protein that undergoes cleavage upon mitochondrial import leading to its proteasomal degradation. Under depolarizing conditions, it accumulates on mitochondria where it becomes activated, phosphorylating both ubiquitin and the ubiquitin E3 ligase Parkin, at Ser65. Our experiments reveal that in retinal pigment epithelial cells, only a fraction of PINK1 becomes stabilized after depolarization by electron transport chain inhibitors. Furthermore, the observed accrual of PINK1 cannot be completely accounted for without an accompanying increase in biosynthesis. We have used a ubiquitylation inhibitor TAK-243 to accumulate cleaved PINK1. Under these conditions, generation of unconjugated "free" phospho-ubiquitin serves as a proxy readout for PINK1 activity. This has enabled us to find a preconditioning phenomenon, whereby an initial depolarizing treatment leaves a residual pool of active PINK1 that remains competent to seed the activation of nascent cleaved PINK1 following a 16-hour recovery period.
Figures

References
-
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K., PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 (2010). - PMC - PubMed
-
- Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H. I., Campbell D. G., Gourlay R., Burchell L., Walden H., Macartney T. J., Deak M., Knebel A., Alessi D. R., Muqit M. M., PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

