TFIIH kinase CDK7 drives cell proliferation through a common core transcription factor network
- PMID: 40020069
- PMCID: PMC11870056
- DOI: 10.1126/sciadv.adr9660
TFIIH kinase CDK7 drives cell proliferation through a common core transcription factor network
Erratum in
-
Erratum for the Research Article "TFIIH kinase CDK7 drives cell proliferation through a common core transcription factor network" by T. Jones et al.Sci Adv. 2025 Jun 6;11(23):eadz0516. doi: 10.1126/sciadv.adz0516. Epub 2025 Jun 6. Sci Adv. 2025. PMID: 40479077 Free PMC article. No abstract available.
Abstract
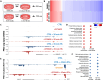
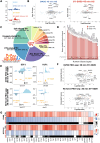
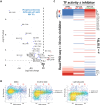
How cyclin-dependent kinase 7 (CDK7) coordinately regulates the cell cycle and RNA polymerase II transcription remains unclear. Here, high-resolution cryo-electron microscopy revealed how two clinically relevant inhibitors block CDK7 function. In cells, CDK7 inhibition rapidly suppressed transcription, but constitutively active genes were disproportionately affected versus stimulus-responsive. Distinct transcription factors (TFs) regulate constitutive versus stimulus-responsive genes. Accordingly, stimulus-responsive TFs were refractory to CDK7 inhibition whereas constitutively active "core" TFs were repressed. Core TFs (n = 78) are predominantly promoter associated and control cell cycle and proliferative gene expression programs across cell types. Mechanistically, rapid suppression of core TF function can occur through CDK7-dependent phosphorylation changes in core TFs and RB1. Moreover, CDK7 inhibition depleted core TF protein levels within hours, consistent with durable target gene suppression. Thus, a major but unappreciated biological function for CDK7 is regulation of a TF cohort that drives proliferation, revealing an apparent universal mechanism by which CDK7 coordinates RNAPII transcription with cell cycle CDK regulation.
Figures



References
-
- Matthews H. K., Bertoli C., de Bruin R. A. M., Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 23, 74–88 (2022). - PubMed
-
- Rimel J. K., Poss Z. C., Erickson B., Hamman K. B., Hu S., Ebmeier C. C., Johnson J. L., Marineau J. J., Carulli J. P., Geyer M., White P., Breault M., Tao L., DeRoy P., Clavet C., Nayak S., Iwasa J., Bentley D. L., Dowell R. D., Old W. M., Taatjes D. J., Selective inhibition of CDK7 reveals high-confidence targets and new models for TFIIH function in transcription. Genes Dev. 34, 1452–1473 (2020). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

