Characterization of challenging forensic DNA traces using advanced molecular technologies
- PMID: 40021558
- PMCID: PMC12170681
- DOI: 10.1007/s00414-025-03448-8
Characterization of challenging forensic DNA traces using advanced molecular technologies
Abstract
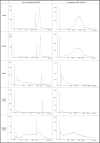
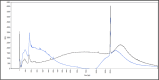
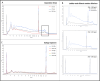
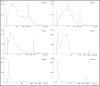
The majority of crime scenes contain DNA that is either present in small amounts or degraded, making it difficult to obtain usable DNA profiles using conventional technologies. The current standard for analyzing casework samples is the specific amplification of short tandem repeats (STR), which is limited by DNA quality and quantity. Since the goal of forensic science is to identify a suspect or victim regardless of trace quality, we evaluated three technological approaches to better characterize and exploit these traces: (i) ultra-sensitive pulse-field electrophoresis on a Femto Pulse System (FPS) to visualize DNA content, (ii) real-time quantitative PCR based on Alu repeats to quantify human DNA and analyze its integrity, and (iii) 16S ribosomal RNA gene (16S rRNA) amplicon sequencing to identify microbiota. We optimized FPS analysis using DNA from model traces (blood, saliva, semen, touch DNA, and vaginal swabs) and applied the protocol to 100 casework samples. We found differences between the FPS profiles of model and casework samples, showing a variation in fragment size and distribution, suggesting the presence of non-human DNA. Using Alu-qPCR and 16S rRNA amplicon sequencing, we determined the amount and proportion of human and non-human DNA. Human DNA was detected in 84% of traces with an average of 70 pg per trace, while 16S rRNA revealed microbial DNA as the most abundant DNA in traces. These analyses provide new insights into forensic trace composition, allowing better sorting and profiling of traces.
Keywords: Alu sequences; DNA trace; Degraded DNA; Femto pulse system; Forensic casework; Microorganism analysis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Approval was obtained from the ethics committee of the Judicial Center of the National Gendarmerie. The procedures used in this study adhere to the tenets of the French National Charter for research integrity (2015). Consent to participate: Informed consent was obtained from all individual participants included in the study. Research involving human participants: All human participants are in agreement with this study. Competing interests: The authors have no competing interests to declare and nothing to disclose. Declaration of generative AI and AI-assisted technologies in the writing process: No tool used.
Figures



References
-
- (2016) Buckleton JS, Bright JA, Taylor D, editors. Forensic DNA Evidence Interpretation. 2nd ed. Boca Raton: CRC Press. 508. 10.4324/9781315371115
-
- Virkler K, Lednev IK (2009) Analysis of body fluids for forensic purposes: From laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int. 10.1016/j.forsciint.2009.02.013 - PubMed
-
- Hall CL, Zascavage RR, Sedlazeck FJ, Planz JV (2020Jan) Potential applications of nanopore sequencing for forensic analysis. Forensic Sci Rev 32:23–54 - PubMed
-
- Hall A, Sims LM, Ballantyne J (2014) Assessment of DNA damage induced by terrestrial UV irradiation of dried bloodstains: Forensic implications. Forensic Sci Int Genet. 10.1016/j.fsigen.2013.06.010 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

