Transcriptional diversification in a human-adapting zoonotic pathogen drives niche-specific evolution
- PMID: 40021638
- PMCID: PMC11871327
- DOI: 10.1038/s41467-025-57331-6
Transcriptional diversification in a human-adapting zoonotic pathogen drives niche-specific evolution
Abstract
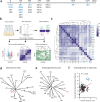
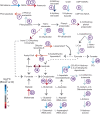
Bacterial pathogens can undergo striking adaptive evolutionary change in the context of infection, driven by selection forces associated with host defenses and antibiotic treatment. In this work, we analyze the transcriptional landscape associated with adaptation in an emerging zoonotic pathogen, Bordetella hinzii, as it evolved during a 45-month infection in an IL12Rβ1-deficient immunocompromised host. We find evidence of multiple niche-specific modifications in the intravascular and gastrointestinal compartments, involving the superoxide dismutase system, glutamate and ectoine metabolism, chaperone-mediated protein folding, pilus organization, and peptide transport. Individual blood lineages displayed modifications in glutathione, phenylacetate, and 3-phenylpropionate metabolism, iron cluster assembly, and electron transport, whereas individual gastrointestinal lineages demonstrated changes relating to gluconeogenesis, de novo pyrimidine synthesis, and transport of peptides and phosphate ions. Down regulation of the flagellar operon with corresponding loss of flagellar structures occurred in multiple lineages, suggesting an evolutionary tradeoff between motility and host immune evasion. Finally, methylome analysis demonstrates alteration of global genome methylation associated with loss of a Type III methyltransferase. Our findings reveal striking plasticity in how pathogen transcriptomes explore functional space as they evolve in the context of host infection, and demonstrate that such analysis may uncover phenotypic adaptations not apparent from genomic analysis alone.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Dekker, J. P. Within-host evolution of bacterial pathogens in acute and chronic infection. Annu Rev. Pathol.19, 203–226 (2024). - PubMed
-
- Barreto, H. C. & Gordo, I. Intrahost evolution of the gut microbiota. Nat. Rev. Microbiol21, 590–603 (2023). - PubMed
-
- Smith, W. P. J., Wucher, B. R., Nadell, C. D. & Foster, K. R. Bacterial defences: mechanisms, evolution and antimicrobial resistance. Nat. Rev. Microbiol21, 519–534 (2023). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

