HBV-associated hepatocellular carcinomas inhibit antitumor CD8+ T cell via the long noncoding RNA HDAC2-AS2
- PMID: 40021665
- PMCID: PMC11871238
- DOI: 10.1038/s41467-025-57367-8
HBV-associated hepatocellular carcinomas inhibit antitumor CD8+ T cell via the long noncoding RNA HDAC2-AS2
Abstract
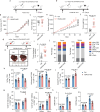
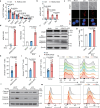
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide. Extracellular vesicles (EV) are critical mediators of intercellular communication within the tumor microenvironment, and cancer-cell-secreted EVs often facilitate cancer progression. Here we show that in HBV-associated HCC, tumor-cell-derived EVs contain a TGFβ-inducible long noncoding RNA, termed HDAC2-AS2. EVs enriched with HDAC2-AS2 facilitate cancer progression by suppressing cytotoxicity of intra-tumor CD8+ T cells. Mechanistically, in activated cytotoxic CD8+ T cells, translocation of the transcription factor cyclin-dependent kinase 9 (CDK9), to the cytoplasm is critical for functional integrity. HDAC2-AS2 targets and blocks cytosolic CDK9, and this results in exhaustion of PD-1+CD8+ T cells and suppression of IFN-γ+CD8+ T cell cytotoxicity. Notably, we demonstrate that low CDK9 and high HDAC2-AS2 expressions are associated with poor survival of HCC, which can be rescued by anti-PD-1 therapy. These findings emphasize the significance of tumor-derived EVs in suppressing antitumor CD8+ T cell immunity to promote tumorigenesis, and highlight extracellular HDAC2-AS2 as a promising biomarker and therapeutic target for HCC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Forner, A., Reig, M. & Bruix, J. Hepatocellular carcinoma. Lancet391, 1301–1314 (2018). - PubMed
-
- Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim.7, 6 (2021). - PubMed
-
- Yoh, T. et al. Surgery for recurrent hepatocellular carcinoma: achieving long-term survival. Ann. Surg.273, 792–799 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

